Humans have always been fascinated with reverse engineering, whether to create Frankenstein or artificial organs. This science fiction is slowly becoming a reality using organ-on-chip and tissue engineering technology. In organ-on-chip technology, the physiological function of a human organ is closely mimicked inside a microfluidic channel. Tissue engineering may involve bio-printing of living cells inside a scaffold to mimic a whole organ. Both these technologies offer great promise for developing personalized medicine and studying disease models. The successful use of this technology will depend on the ability to maintain healthy cell environment and monitor biological processes inside microfluidic channels for an extended period of time (~2-4 weeks). By integrating sensors inside microfluidic channels, these parameters can be continuously monitored. The advantages of integrating sensors into microfluidic channels are:
- Label-free continuous monitoring of cell health inside channels
- Low volumes (picoliters) in microfluidic channels enable high sensitivity of detection
- By placing sensors close to the cells, the analyte of interest is less likely to be diluted in cell media
Truly mimicking human physiology
Living cells are sensitive even to the slightest changes in their environment and release certain molecules in response to these changes. Monitoring these molecules provides valuable information on the state of health and can predict response to drugs. For example, Zhang et al.1 developed a multi-sensor integrated organ-on-chip platform to monitor micro-environmental parameters such as pH, oxygen, and temperature by using optical-based sensors. Additionally, the authors developed label-free electrochemical immunobiosensors for continuous monitoring of molecules secreted by tissue-engineered organs inside the microfluidic channels. This work demonstrated that organ-on-chips can integrate real-time monitoring of the biophysical and biochemical parameters in a micro-environment.
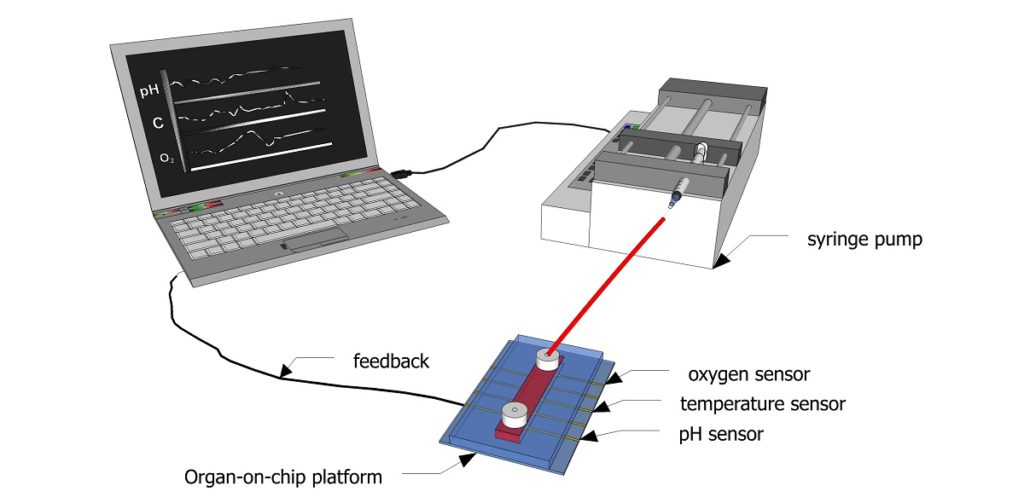
Figure 1: Schematic showing how sensors can be used to continuously monitor biochemical parameters inside a microfluidic channel.
Credit: Aditya Aryasomayajula
Figure 1 shows a schematic diagram of how feedback from the sensors integrated into a microfluidic channel can be used for regulating the microenvironment for cell culture. Sensors can continuously monitor biophysical and biochemical parameters inside a channel and directly communicate this information to a computer, which in turn adjusts the inflow of fresh cell culture media using a pump. Thismicroperfusion of the channel with cell culture media can help maintain an optimal pH, oxygen level, and temperature. This feedback can be used to improve cell viability for organ-on-chip applications and bring us a step closer to truly mimicking the human physiology.
Currently, there are developed sensors for measurement of pH2, oxygen3, glucose3 and lactate4. Integrating any combination of these sensors into organ-on-chip devices will help define the microenvironment that may be important in drug screening applications.
Challenges integrating sensors and outlook
There are several challenges that one has to overcome in order to successfully integrate sensors into microfluidic channels. They are as follows:
a. Biofouling
One of the major challenges of using sensors for long-term monitoring is biofouling of the surface. Biofouling refers to the forming of a thin biofilm on the surface of the sensing material which results in degrading the performance of the sensors. This problem is particularly amplified when sensing in biological environments. There have been several strategies to mitigate biofouling of sensors. For example, surface modification (silanization) is a popular technique to extend the life of sensors.
b. Fabrication technologies
The conventional use of soft lithography for fabricating microfluidic devices requires modifications to integrate sensors directly into the channels. Problems, such as bonding of PDMS surface and aligning sensors inside channels, may be a challenge. New fabrication techniques like LEGO and cartridge assemblies have gained popularity for manufacturing organ-on-chip devices, as these are better suited for sensor integration.
c. Sensor sensitivity and selectivity
Sensor sensitivity refers to the detection limit of the sensors. Ultrasensitive sensors are required for sensing in low volumes (picoliters) in the mg/L or ng/L range. Recently nanomaterials like graphene5 and carbon nanotubes6 have gained popularity because of their high sensitivity to detect in low volumes. Selectivity is another major challenge for these sensors. This parameter refers to the ability of the sensors to detect only specific molecule of interest with high signal to noise ratio. The presence of various interfering species in cell culture media can be a challenge to sense specifically the molecule of interest. Aptamer-based sensors offer good selectivity with high signal to noise ratio.
In conclusion, new technology brings new demands and challenges. Organ-on-chip and tissue engineering show great promise for personalized medicine, drug development, and studying disease models with more complex physiologically relevant systems. Integrating sensors into microfluidic channels can enhance the performance of these technologies by continuous monitoring of biophysical and biochemical parameters in micro-environments.
1. Zhang, Y.S., Aleman, J., Shin, S.R., Kilic, T., Kim, D., Shaegh, S.A.M., Massa, S., Riahi, R., Chae, S., Hu, N. and Avci, H., 2017. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proceedings of the National Academy of Sciences, p.201612906.
2. Welch, D. and Christen, J.B., 2014. Real-time feedback control of pH within microfluidics using integrated sensing and actuation. Lab on a Chip, 14(6), pp.1191-1197.
3. Rodrigues, N.P., Sakai, Y. and Fujii, T., 2008. Cell-based microfluidic biochip for the electrochemical real-time monitoring of glucose and oxygen. Sensors and Actuators B: Chemical, 132(2), pp.608-613.
4. Weltin, A., Slotwinski, K., Kieninger, J., Moser, I., Jobst, G., Wego, M., Ehret, R. and Urban, G.A., 2014. Cell culture monitoring for drug screening and cancer research: a transparent, microfluidic, multi-sensor microsystem. Lab on a Chip, 14(1), pp.138-146.
5. Shao, Y., Wang, J., Wu, H., Liu, J., Aksay, I.A. and Lin, Y., 2010. Graphene based electrochemical sensors and biosensors: a review. Electroanalysis, 22(10), pp.1027-1036.
6. Jacobs, C.B., Peairs, M.J. and Venton, B.J., 2010. Carbon nanotube based electrochemical sensors for biomolecules. Analytica chimica acta, 662(2), pp.105-127.
Enjoyed this article? Don’t forget to share.

Aditya Aryasomayajula
Aditya currently holds a joint postdoctoral fellowship in Mechanical Engineering Department at McMaster University and Firestone Institute for Respiratory Health at St. Joseph’s Healthcare, Hamilton. He is also the project manager for the sensor division of Global Water Futures program. He has won prestigious scholarships from National Science Foundation (NSF) and Deutsche Forschungsgemeinschaft (DFG) Germany. His research interests include developing sensors for environmental monitoring and healthcare diagnostics. He is currently developing a lung-on-chip device to study disease models like asthma and drug screening for cystic fibrosis.