Sequencing has created a lot of buzz in the last few decades and now is a big industry with many commercially available machines that can sequence genes ranging from few to billions of bases. On the other hand, several microfluidic platforms have been developed to improve the sequencing technology1,2 ranging from sample enrichment to the final read of the gene sequence, with many of them being commercialized. Sample enrichment is one of the major requirements for sequencing, its efficiency is very important which can be achieved by microfluidic approaches1. This article emphasizes that microfluidics is vital for the development of advanced DNA sequencing techniques, for instance, Targeted-Next Generation Sequencing (NGS).
DNA sequencing – determining the order of the four building blocks (nucleotide bases) that make up the DNA strand is not only useful in studying fundamental biological processes but also in applied fields such as diagnostic. By comparing sequencing results to reference genomes, researchers can identify genetic variations which can cause various diseases or disorders like cancer, Johanson-Blizzard syndrome3 etc. Moreover, the study of these genetic variations enabled by NGS is the backbone for the development of personalized medicine. NGS can sequence nucleotides faster and at much cheaper rates compared to Sanger sequencing(traditional) method. To further reduce the cost and increase the sequencing depth only certain genes of interest can be sequenced. These specific genes are the protein-coding sections and are known toharbor mutations. With “targeted NGS” such regions can be sequenced instead of having to sequence the whole genome4. Before targeted NGS is performed, the genes of interest need to be isolated and enriched from the rest of the genome with high specificity and sensitivity. Herein, comes the role microfluidics which can substitute the cumbersome labor-intensive target enrichment methods.
Microfluidicswhich enables the manipulation of liquid at extremely small volumes and particularly droplet microfluidics has gained wide popularity in the development of different aspects of NGS5. Amongst other areas of NGS development, several efforts have also been made where microfluidics technology is used to isolate genes of interest from the rest of the genome and enrich them for targeted sequencing. Some of the techniques have shown promising outcomes and have been adopted and integrated into commercial products.

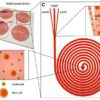
Figure 1: The work was done by Eastburn et al. – Droplet-based microfluidic system for target gene enrichment. The workflow shows the encapsulation of the target gene and at the end sorted and separated from other droplets. Credit: NCBI
RainDance technologies developed a platform – ThunderStormthat conducts polymerase chain reaction (PCR) inside millions of microdroplets in parallel. The platform uses a microfluidic device that can generate and merge the droplets yielding a high target gene concentration. Fluidigm’s microfluidic Access Array platform can prepare enriched libraries for NGS, compatible with several sequencing machines. This platform was used in a study to perform parallel amplification using several primers and screening of 5995 genomic bases was done to identify cancer mutations6. A more recent advancement to isolate target gene using PCR was demonstrated Eastburn et al. In this work the authors presented a novel approach to encapsulate DNA molecules into millions of microdroplets, followed by TaqMan PCR reactions to identify the droplets containing the target gene. Based on the fluorescence signal, the drops containing the target gene were sorted (Figure 1). After the isolation and enrichment of the target gene, it is now ready to undergo sequencing. This method has shown to overcome the specificity issue which is common to the other target gene isolation techniques7.
Hybridization is another approach to enrich target sequences and is known for its ability to maintain uniformity with the encapsulation of target genes. Furthermore, this method of target enrichment can be coupled with the present high throughput NGS technology. The Geniom Biochip uses microfluidic approach to enrich target DNA in the sample using this principle of hybridization. Using this platform, a new strategy was developed and reported to have an efficient capture of the target genes with higher depth. To do so two iterative cycles of hybridization were performed with the microfluidic platform and the enriched target gene was then sequenced for cancer-related mutations8.
The role of microfluidics mentioned here highlights only one among many other areas where it is helping in the development of NGS. With the expeditious development of NGS utilizing microfluidics in the backdrop, it is expected that microfluidics will continue to play many such significant roles in the future. Apart from the development and advancements of different microfluidic platforms for NGS, more collaborations from different disciplines are essential for more significant future breakthroughs and commercialization in the field of sequencing.
1. Tewhey, Ryan, et al. “Microdroplet-based PCR enrichment for large-scale targeted sequencing.” Nature biotechnology 27.11 (2009): 1025.
2. Kartalov, Emil P., and Stephen R. Quake. “Microfluidic device reads up to four consecutive base pairs in DNA sequencing‐by‐synthesis.” Nucleic acids research 32.9 (2004): 2873-2879.
3. Zenker, Martin, et al. “Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome).” Nature genetics 37.12 (2005): 1345.
4. Carter, Tonia C., and Max M. He. “Challenges of identifying clinically actionable genetic variants for precision medicine.” Journal of healthcare engineering 2016 (2016).
5. Ma, Sai, Travis W. Murphy, and Chang Lu. “Microfluidics for genome-wide studies involving next generation sequencing.” Biomicrofluidics 11.2 (2017): 021501.
6. Forshew, Tim, et al. “Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA.” Science translational medicine 4.136 (2012): 136ra68-136ra68.
7. Eastburn, Dennis J., et al. “Microfluidic droplet enrichment for targeted sequencing.” Nucleic acids research 43.13 (2015): e86-e86.
8. Summerer, Daniel, et al. “Microarray-based multicycle-enrichment of genomic subsets for targeted next-generation sequencing.” Genome research 19.9 (2009): 1616-1621.
Enjoyed this article? Don’t forget to share.

Shubhodeep Paul
Shubhodeep Paul is currently a Ph.D. student at the University of Texas at Arlington majoring in Mechanical Engineering. He received his BS in Mechanical Engineering from West Bengal University of Technology, India (2013). His current research is focused on microfluidics-based purification and sample preparation techniques for different lab-on-a-chip applications. Shubhodeep also has a keen interest in molecular biology, organ-on-a-chip engineering, and synthetic biology.