In conventional laboratories, a range of available technologies enables scientists to genetically engineer cells, study their migration patterns, determine their mechanical properties and even analyze genetic differences. Nevertheless, protocols for such experiments are set in place by standard equipment and commercial kits, often requiring considerable labor and cost. Although some degree of flexibility is permitted in the alteration of certain parameters, results could be compromised by such change. As such, most biologists are typically averse to making changes to their setup. Performing experiments has thus far meant using gold-standard instruments from established pharmaceutical and biotechnological companies. For example, one could start their day preparing 10 L of media for culturing microorganisms in a large chemostat; use a DNA extraction kit to lyse the cells, numerous rounds of centrifugation; DNA purification; proceed on to amplification and target detection using a benchtop real-time Polymerase Chain Amplification (rt-PCR) machine. However, consider this. What if a device the size of a standard laboratory glass slide is able to accomplish these steps from cell culture to mutation identification in half the time and perhaps even half the cost?
Microfluidics in Biology
Microfluidics is one such technology that has demonstrated great potential in reducing the time and cost of conventional laboratory assays. Briefly, microfluidics is the control of fluids in channels at the scale of tens to hundreds of micrometer. As a result of the small scale at which the fluids are processed, the flow is typically laminar (Reynolds number <100) and can be controlled very precisely. Given that most biological samples contain thousands to millions of cells to be analyzed and can be relatively hard to collect, microfluidics is especially suited for biological work as it allows for high throughput processing with only a small amount of sample.
The idea of Lab-On-a-Chip (LOC) using microfluidic technology is to integrate current and novel research principles into a single miniaturized device in the nano- to micro-scale. However, it is noteworthy that the idea of microfluidics is not entirely recent. Fluid manipulation in microchannels has been around since the 1950s. However, the field centered around devices fabricated using integrated circuit technology limited to silicon substrates, while taking advantage of the electroosmotic flow to manipulate their targets. With the emergence of softer materials and advanced fabrication techniques in the 1990s, microfluidics gained widespread popularity amongst biologists and engineers1. From cell-based assays2, genomics3, proteomics4,5, transcriptomics5 and even drug discovery6, microfluidics have branched out in many disciplines of biological sciences. Microfluidic technology can be further categorized into two classes, employing either active sorting or passive (label-free) sorting methods to control targets. With the advancement in microfluidics, the analyses of individual cells are now a possibility.
1. The Need for Single Cells
In many scientific studies, bulk data are often used as a representation to interpret results. However, the major drawback of this technique is the possible heterogeneity within populations. One well-researched example is circulating tumour cells (CTCs) present in the blood of metastatic cancer patients. CTCs are tumor cells that disseminate into the bloodstream from the primary tumor to begin the metastasis process. Although most tumour cells in circulation do not survive the hostile immune environment and high shear experienced in blood vessels, some CTCs do successfully travel and establish at a distant site to form a metastatic lesion. While whole blood can be obtained using liquid biopsy, a non-invasive technique, the extremely low numbers of CTCs (1-10/mL) would mean taking the average from whole blood would likely arrive at results biased towards wild-type readings such as white blood cells. Moreover, due to the low numbers of these cells, the genetic information may also be masked. Microfluidics has enabled the fractionation and isolation of CTCs. Through this, genomic analysis can be conducted at the individual cell level which can reveal critical information about the mutational status of the cancer patient. This would then allow clinicians to personalize treatment and improve the survival in these patients.
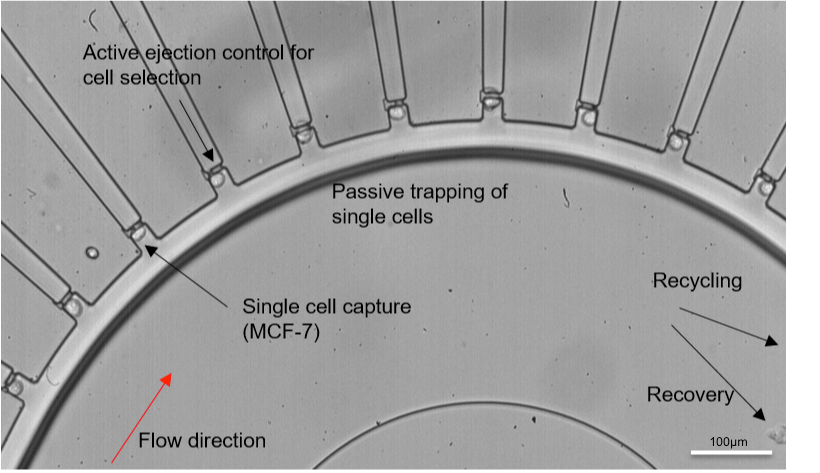
Figure 1. A microfluidic single cell isolation device. Image adapted and reproduced with permission from Yeo et al., Sci. Rep. 6, 22076 (2016). Copyright Nature Publishing Group.
2. Portability
In developing countries, infectious diseases are a widespread problem for people and economic development. This situation is worsened by a lack of well-equipped diagnostic laboratories. The usual detection by molecular assays such as the enzyme-linked immunosorbent assay, western blotting and PCR requires skilled technicians and several hours of processing. Therefore, a potential solution is to have a portable point-of-care (POC) device providing rapid detection. Particularly, one emerging technique, involving the use of paper microfluidics, has shown promise in the detection of pathogens such as HIV with minimal steps7. Paper microfluidics have also been deployed successfully in the field to detect Ebola8. The low fabrication cost, diverse capabilities and small size of microfluidic devices make them suitable for POC diagnostics. In particular, paper microfluidics eliminates the need for a laboratory and its bulky equipment in order to perform these tests.
Outlook
Globally, labs are pushing the boundaries of engineering and biology. Aside from its apparent portable size, additional advantages of microfluidics are as follows:
- Flexibility in manipulation
- Lower reagent
- Affordability
- Real-time monitoring
There are other numerous physical phenomena used in fluid dynamics. Aside from paper-based methods, there are also the following methods which are commonly used,
- Droplet microfluidics
- Digital microfluidics
- Sensor microfluidics
- Optofluidics
With countries transitioning from basic to translational research, the integration and collaborations across different expertise are pivotal. Nevertheless, despite the increasing numbers of microfluidic-related publications, few devices end up being commercialized. One barrier to this is the automation of microfluidic devices requires shrinking of concomitant components. Another pressing issue is a need to prevent ephemeral graduate projects by encouraging aspiring scientists to collaborate with industries and clinicians and identify a market need and to go further by developing the device for commercial use. The pressure of looking for a novel method for publication means students spending years working on a design from conception instead of developing and optimizing a pre-existing one.
That being said, the diversity of research in microfluidics can also generate better ideas and competition may drive breakthrough projects. As compared to conventional lab-bench equipment, the higher throughput seen in microfluidics means obtaining results in a shorter time coupled with higher spatial and temporal resolution. Already, with the advancement in microfluidics, we are moving towards a minimally invasive tool for cancer assessment and detecting pathogens in resource-limited areas. Hopefully, in the near future, we will develop a POC device which can provide a rapid health analysis from the comfort of our homes without the need for long waits at the hospital. Microfluidics is revolutionizing the way experiments are performed by transferring the diagnostic capabilities from lab bench to consumer’s hands, and it may continue to complement research in science in unprecedented ways.
1. Duffy, D.C., et al., Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane). Anal Chem, 1998. 70(23): p. 4974-84.
2. Tumarkin, E., et al., High-throughput combinatorial cell co-culture using microfluidics. Integr Biol (Camb), 2011. 3(6): p. 653-62.
3. Kim, S., et al., High-throughput automated microfluidic sample preparation for accurate microbial genomics. Nature Communications, 2017. 8: p. 13919.
4. Abdel-Sayed, P., et al., Fabrication of an Open Microfluidic Device for Immunoblotting. Analytical Chemistry, 2017. 89(18): p. 9643-9648.
5. Genshaft, A.S., et al., Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biology, 2016. 17(1): p. 188.
6. Tsui, J.H., et al., Microfluidics-assisted in vitro drug screening and carrier production. Advanced Drug Delivery Reviews, 2013. 65(11): p. 1575-1588.
7. Rohrman, B.A. and R.R. Richards-Kortum, A paper and plastic device for performing recombinase polymerase amplification of HIV DNA. Lab on a Chip, 2012. 12(17): p. 3082-3088.
8. Magro, L., et al., Paper-based RNA detection and multiplexed analysis for Ebola virus diagnostics. Scientific Reports, 2017. 7(1): p. 1347.
Enjoyed this article? Don’t forget to share.

Trifanny Yeo
Trifanny Yeo is currently a research assistant at the Department of Biomedical Engineering, National University of Singapore. She received her BSc from the University of Queensland and has been working with microfluidics, developing devices for use in personalized medicine. Her research interest also includes transcriptomics, nanomaterials and wearable electronics.