04 Aug Transcriptome-based profiling using droplet microfluidics
Abstract
“Anti-cancer therapies often exhibit only short-term effects. Tumors typically develop drug resistance causing relapses that might be tackled with drug combinations. Identification of the right combination is challenging and would benefit from high-content, high-throughput combinatorial screens directly on patient biopsies. However, such screens require a large amount of material, normally not available from patients. To address these challenges, we present a scalable microfluidic workflow, called Combi-Seq, to screen hundreds of drug combinations in picoliter-size droplets using transcriptome changes as a readout for drug effects. We devise a deterministic combinatorial DNA barcoding approach to encode treatment conditions, enabling the gene expression-based readout of drug effects in a highly multiplexed fashion. We apply Combi-Seq to screen the effect of 420 drug combinations on the transcriptome of K562 cells using only ~250 single cell droplets per condition, to successfully predict synergistic and antagonistic drug pairs, as well as their pathway activities.”

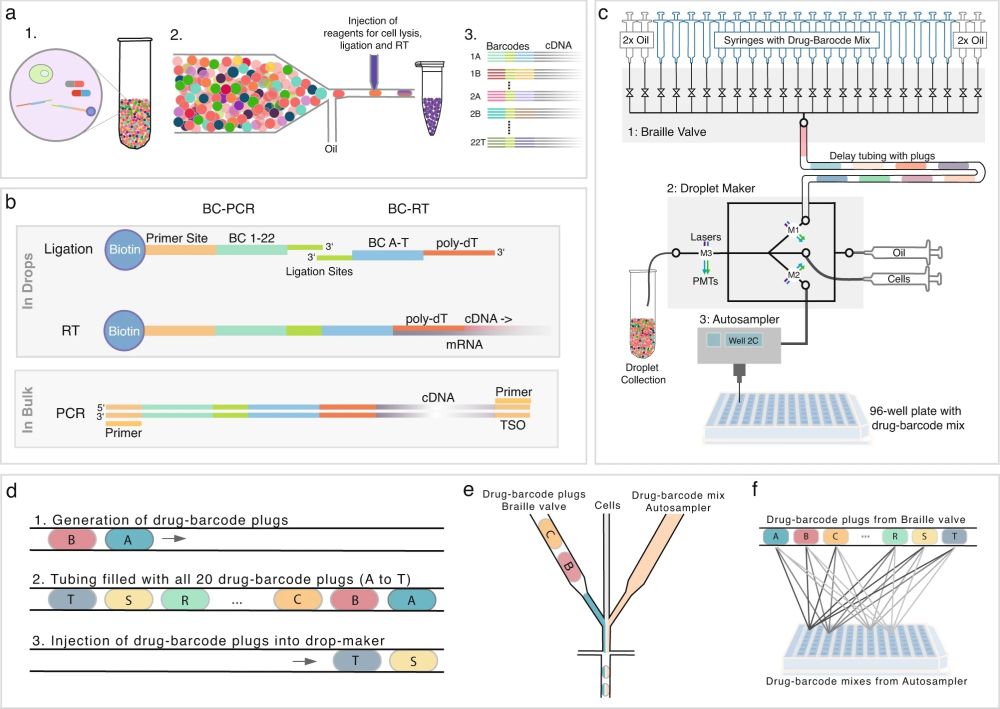
a Overview Combi-Seq workflow: (1) Cells were encapsulated with drug combinations and pairs of barcode fragments encoding drugs. (2) After off-chip incubation, droplets were reinjected into a chip for picoinjection to add reagents for barcode ligation and reverse transcription (RT), enabling barcoding of the transcriptome according to the drug treatment. (3) Upon droplet breakage, pooled libraries for sequencing were generated in which cells from the same treatment group share the same barcode. This facilitated demultiplexing of drug treatments and gene expression-based readouts for pools of 250 cells. b Barcoding strategy applied to encode and decode drug combinations for low input cell pools. Pairs of barcoded PCR primers (BC-PCR) and barcoded poly-dT primers (BC-RT) were joined in a ligation reaction to form functional barcodes, which were used for reverse transcription, thereby encoding a combination of two drugs. By breaking droplets, barcoded cDNA can be recovered and amplified for sequencing. c Microfluidic pipeline used to generate drug combinations in droplets. (1) Braille valves were used to generate sequences of 20 drug-barcode (BC-RT) plugs within the delay tubing. The delay tubing was connected to a drop maker (2) into which cells and drugs-BC-PCR mixtures from multiwell-plates were injected. Finally, injecting the plugs into the drop-maker by opening two oil valves, droplets containing drug pairs with barcodes and cells were generated. M1–M3 indicate positions where fluorescence signals were acquired. d Generation of drug-barcode plugs in the delay tubing: (1) Plugs spaced out by oil were produced by sequentially opening the corresponding valves, (2) resulting in a sequence of 20 drug-barcode plugs. (3) By opening two oil valves, the plugs in the delay tubing were injected into the droplet-maker chip. e Droplet production from a cell suspension, drug-barcode plugs, and drug-barcode mixtures injected by the autosampler, resulting in the co-encapsulation of cells with drug-barcode combinations. f Scheme illustrating the generation of drug combinations from 20 drugs from the valve module with drugs from a 96-well plate. Each sequence of 20 drug-barcode (BC-RT) plugs was combined with drug-barcode (BC-PCR) mixes from one well.” Reproduced under Creative Commons Attribution 4.0 International License from Mathur, L., Szalai, B., Du, N.H. et al. Combi-seq for multiplexed transcriptome-based profiling of drug combinations using deterministic barcoding in single-cell droplets. Nat Commun 13, 4450 (2022).
Figures and the abstract are reproduced from Mathur, L., Szalai, B., Du, N.H. et al. Combi-seq for multiplexed transcriptome-based profiling of drug combinations using deterministic barcoding in single-cell droplets. Nat Commun 13, 4450 (2022). https://doi.org/10.1038/s41467-022-32197-0
Read the original article: Combi-seq for multiplexed transcriptome-based profiling of drug combinations using deterministic barcoding in single-cell droplets


