19 Sep Microfluidics enables high-throughput analysis of the effect of the local environment on gene expression
The spatial organization and cellular microenvironment of the cells can be very important when it comes to RNA sequencing. Therefore, preserving the spatial organization of the cells and monitoring the changes in the cellular surrounding can be crucial for transcriptomic profiling. Droplet-based microfluidics is an ideal candidate for this purpose. For this week’s research highlight, we selected a droplet microfluidics platform that exactly does this! The proposed microfluidic chip was employed to compartmentalize semi-bulk cells along with barcoded beads in droplets and isolate the mRNA to observe the effect of the local cellular environment on RNA transcripts.
“We utilized a microfluidic device specifically designed for the experiments to encapsulate both a barcoded bead and a cell aggregate (a semibulk) into a single droplet. Using sbRNA-seq, we firstly analyzed mouse kidney specimens. In the mouse model, we could associate the pathological information with the gene expression information. We validated the results using spatial transcriptome analysis and found them highly consistent. When we applied the sbRNA-seq analysis to the human breast cancer specimens, we identified spatial interactions between a particular population of immune cells and that of cancer-associated fibroblast cells, which were not precisely represented solely by the single-cell analysis. “, the authors explained.

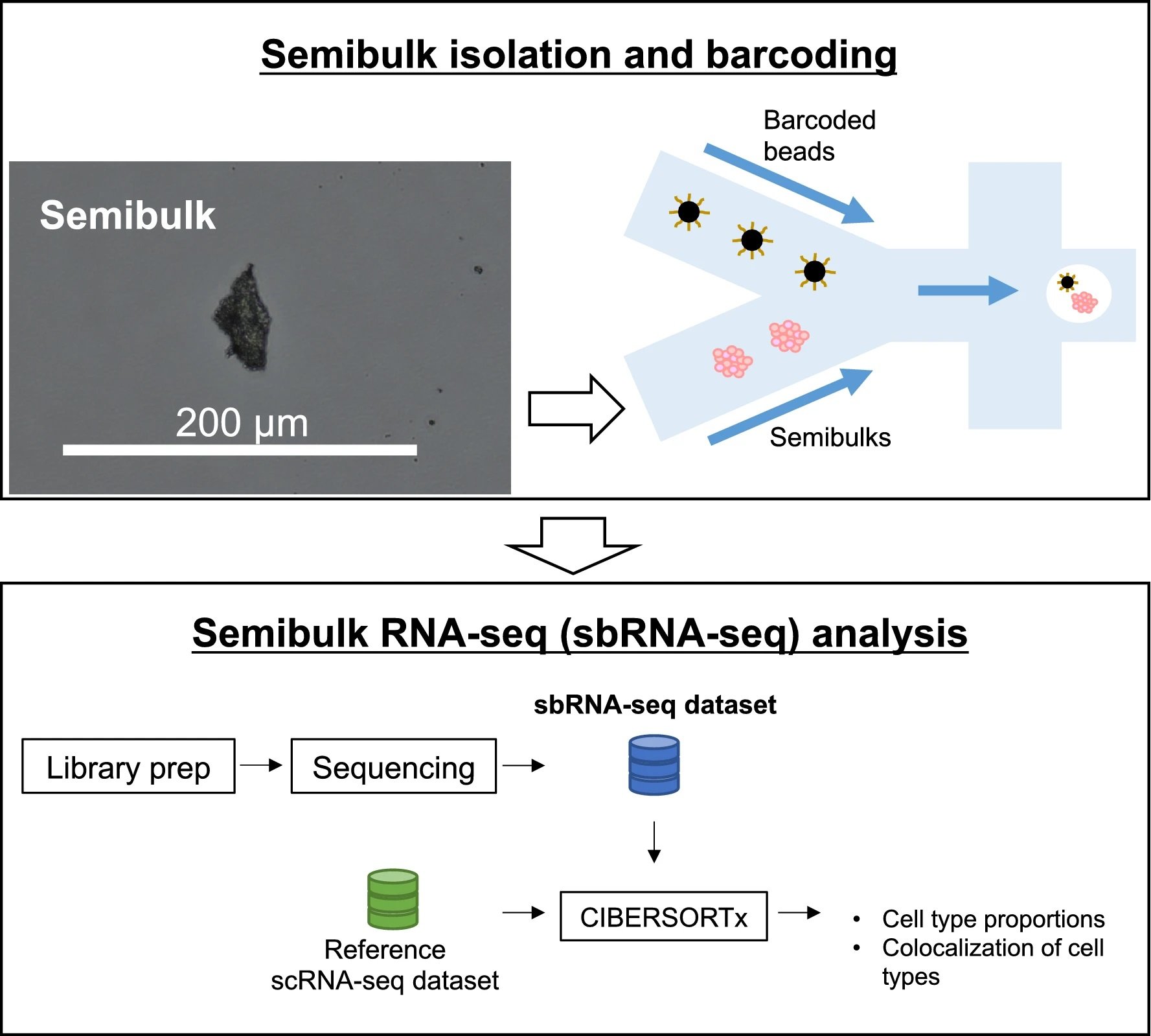
“A workflow of sbRNA-seq. Overall schematic representation. Cell aggregates from tissues are isolated and mixed with a barcoded bead and reaction solution within a droplet. In the droplet, reverse transcription is conducted to generate barcoded cDNA. Library preparation and sequencing are performed to generate sbRNA-seq data. After sequencing, deconvolution analysis is conducted using CIBERSORTx to estimate the cell-type proportion of each semibulk using scRNA-seq data as a reference.” Reproduced under Creative Commons Attribution 4.0 International License from Muto, K., Tsuchiya, I., Kim, S.H. et al. Semibulk RNA-seq analysis as a convenient method for measuring gene expression statuses in a local cellular environment. Sci Rep 12, 15309 (2022).
“we tried to provide a technically easier and more cost-effective platform to analyze the spatial organization of the gene expressions in tissues. Key features that distinguish it from the other platforms include a PDMS microfluidic device designed for semibulks which can be used to analyze several thousands of semibulks in a single experiment. The filtered samples can be directly applied to this system without enzyme, which reduces the damage to mRNAs.“, the authors explained.
The figures and the abstract are reproduced from Muto, K., Tsuchiya, I., Kim, S.H. et al. Semibulk RNA-seq analysis as a convenient method for measuring gene expression statuses in a local cellular environment. Sci Rep 12, 15309 (2022). https://doi.org/10.1038/s41598-022-19391-2 under Attribution 4.0 international (CC BY 4.0) licences.
Read the original article: Semibulk RNA-seq analysis as a convenient method for measuring gene expression statuses in a local cellular environment


