24 Nov Mass spectrometry and microfluidics meet to enable sensing of product formation
“Enzymes are represented across a vast space of protein sequences and structural forms and have activities that far exceed the best chemical catalysts; however, engineering them to have novel or enhanced activity is limited by technologies for sensing product formation. Here, we describe a general and scalable approach for characterizing enzyme activity that uses the metabolism of the host cell as a biosensor by which to infer product formation. Since different products consume different molecules in their synthesis, they perturb host metabolism in unique ways that can be measured by mass spectrometry. This provides a general way by which to sense product formation, to discover unexpected products and map the effects of mutagenesis.”
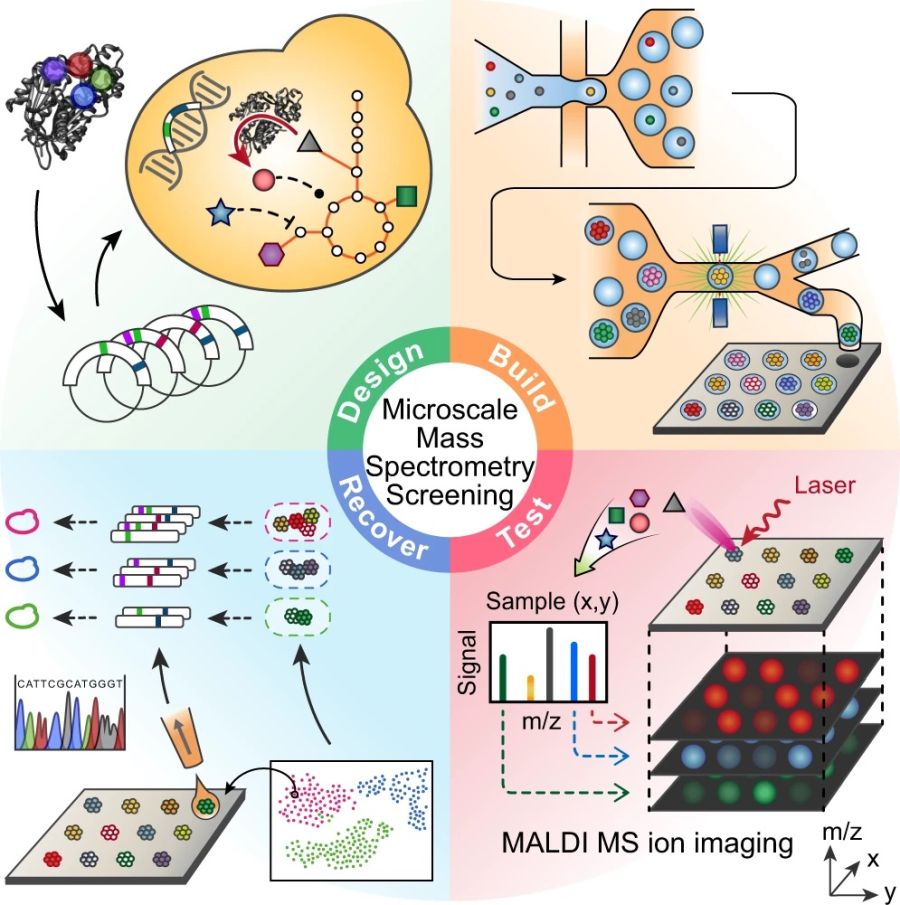
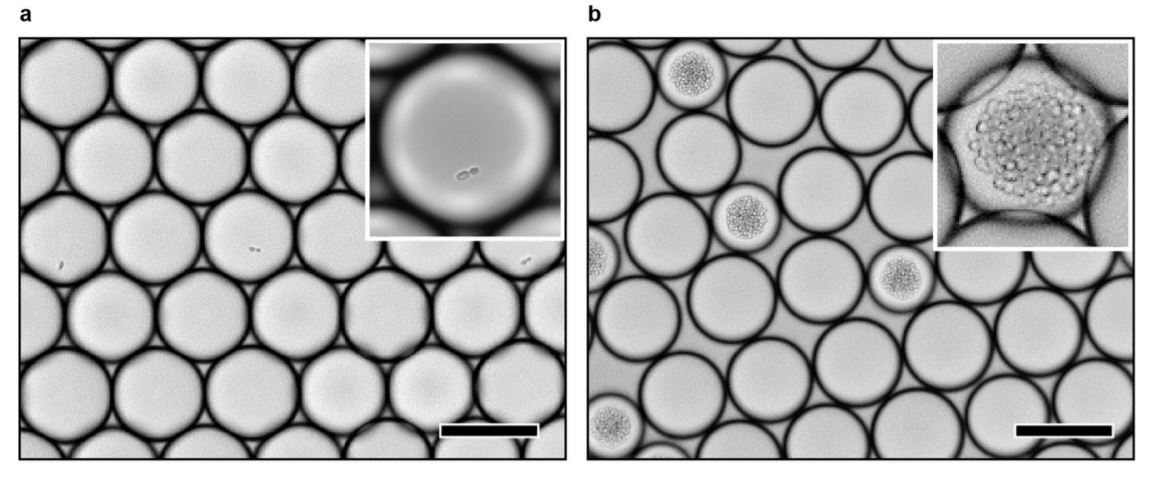
“Enzyme variants are designed and transformed into yeast (design) and then synthesized in the yeast where they consume molecules of central metabolism to generate product (build). Using printed droplet microfluidics, they are dispensed to a picoliter well array and subjected to MALDI-MS imaging to quantify cell metabolites (test). UMAP clusters cells according to metabolomic profile, where each cluster indicates a different enzyme phenotype. Desired mutants are extracted from the plate, sequenced, and confirmed in bulk cultures.” Reproduced under Creative Commons Attribution 4.0 International License from Xu, L., Chang, KC., Payne, E.M. et al. Mapping enzyme catalysis with metabolic biosensing. Nat Commun 12, 6803 (2021).
Figures and the abstract are reproduced from Xu, L., Chang, KC., Payne, E.M. et al. Mapping enzyme catalysis with metabolic biosensing. Nat Commun 12, 6803 (2021). under Creative Commons Attribution 4.0 International License
Read the original article: Mapping enzyme catalysis with metabolic biosensing


