16 Apr Machine learning-aided microfluidic platform predicts the skin-sensitization potential of drugs
The emergence of machine learning techniques to help scientific research has opened new windows of opportunities for researchers and has empowered the research community with unprecedented prediction capabilities. Machine learning techniques are finding their way into the microfluidic domain helping researchers to boost their data acquisition and analysis pipeline. In this week’s research highlight, we will introduce one such microfluidic advancement. In a recent paper published in the Lab on a chip journal, a research team developed a microfluidic chip for multicellular coculture along with machine learning techniques to predict cutaneous drug reactions.
“Here, we report a novel in vitro drug screening platform, which comprises a microfluidic multicellular coculture array (MCA) to model different mechanisms-of-action using a collection of simplistic cellular assays. The resultant readouts are then integrated with a machine-learning algorithm to predict the skin sensitizing potential of systemic drugs. The MCA consists of 4 cell culture compartments connected by diffusion microchannels to enable crosstalk between hepatocytes that generate drug metabolites, antigen-presenting cells (APCs) that detect the immunogenicity of the drug metabolites, and keratinocytes and dermal fibroblasts, which collectively determine drug metabolite-induced FasL-mediated apoptosis.“, the authors explained.

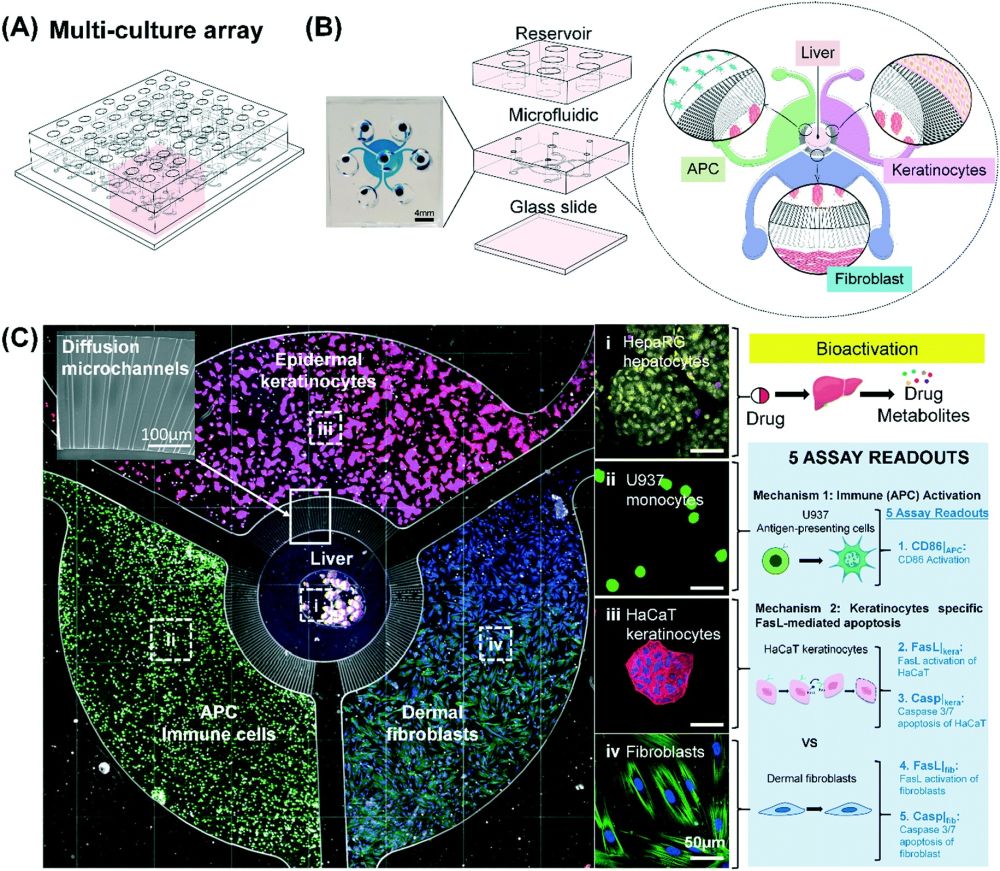
Modeling multiple mechanisms involved in cutaneous drug reactions in a multi-culture array (MCA) platform. (A) Schematic of the MCA showing 9 coculture units in an array format amenable for drug screening. (B) Magnified view of a single coculture unit showing the compartmentalization of liver, immune (APC), keratinocytes and fibroblasts by diffusive microchannels which allow for interactions via soluble factors. (C) Fluorescent images show HepaRG-hepatocytes spheroid (HHS) which are located in the center compartment with the surrounding of HaCaT (pink), human dermal fibroblasts (blue), and U937 (green) in their own annular compartments respectively. Different cell compartments are connected by micro-diffusion channel. The images on the right is a schematic illustrating two possible mechanisms to cause cutaneous drug reactions. Reproduced from L. H. Chong, T. Ching, H. J. Farm, G. Grenci, K. Chiam and Y. Toh, Lab Chip, 2022, Advance Article, DOI:10.1039/D1LC01140E. with permission from Royal Society of Chemistry.
The working mechanism of the microfluidic device
Cutaneous side effects have two known origins. One is an immunogenic response as a result of the activation of antigen-presenting cells and the other is a FasL-mediated keratinocytes apoptosis. Therefore, the proposed microfluidic device was designed to have multiple compartments each pertaining to one aspect of the mechanisms involved in cutaneous adverse effects. The compartments were connected with diffusion microchannels to allow cross-talk between these regions. The multicellular coculture array (MCA) microfluidic chips consisted of two PDMS layers. The microfabrication of the microfluidic chips was done via soft-lithography. Quadruple compartments were placed in the bottom layer which were connected with the aforementioned diffusion microchannels. The middle microchamber housed HepaRG-derived hepatocyte spheroids (HHS) that served as a proxy for human hepatocytes. The metabolites produced in this microchamber can travel through the diffusion microchannels to the surrounding compartments that contained U937 antigen-presenting cells (APCs), HaCaT human keratinocytes and human dermal fibroblasts. Imaging-based techniques were used to investigate the effect of the potential drug on these cells leading to its identification as a sensitizing or non-sensitizing compound. The top layer consisted of seven reservoirs and served as the media stock. The media reservoirs were connected to the bottom layer with through-holes. Nine microfluidic units were placed in an array format to allow parallel analysis.
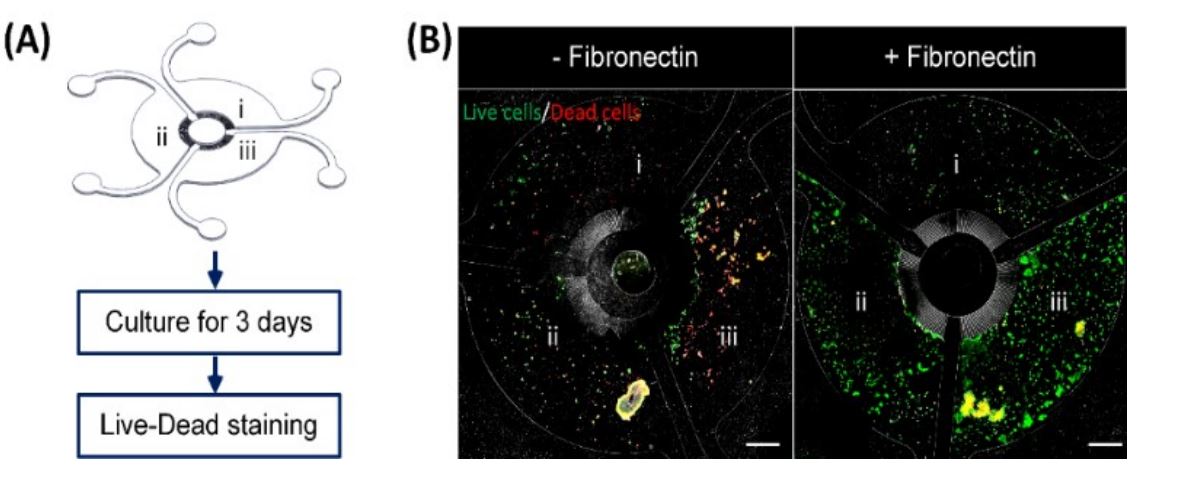
Optimization of cell seeding protocol in MCA. (A) Illustration of the process to optimize the seeding protocol using HaCaT in MCA. Cell suspensions at different cell densities including (i) 0.06 million/mL, (ii) 0.15 million/mL and (iii) 0.3 million/mL of HaCaT were seeded into different compartments of MCA respectively. (B) Fluorescent images of the overall cell seeding in MCA. Live cells are stained with green while dead cells are stained with red. The fibronectin coating (right) is found to promote the cell attachment in the device. Reproduced from L. H. Chong, T. Ching, H. J. Farm, G. Grenci, K. Chiam and Y. Toh, Lab Chip, 2022, Advance Article, DOI:10.1039/D1LC01140E. with permission from Royal Society of Chemistry.
The proposed microfluidic platform was could generate 5 readouts which were then fed into a support vector machine (SVM) to elicit sensitizing or non-sensitizing features of the drugs. The microfluidic devices were tested and validated against 11 FDA-approved drugs. Upon successfully validating the technique, the research team then employed the multicellular culture array and SVM training algorithms to quantify the aforementioned dual mechanism in cutaneous reaction for obeticholic acid (OCA) which has undergone clinical trials to identify the probability of its adverse skin-sensitizing effect.
“The predictive performance of our in vitro model achieves an average of 87.5% accuracy (correct prediction rate), 75% specificity (prediction rate of true negative drugs), and 100% sensitivity (prediction rate of true positive drugs). “, the authors explained.
Figures were reproduced with permission from the Royal Society of Chemistry from L. H. Chong, T. Ching, H. J. Farm, G. Grenci, K. Chiam and Y. Toh, Lab Chip, 2022, Advance Article , DOI: 10.1039/D1LC01140E
Read the original article: Integration of a microfluidic multicellular coculture array with machine learning analysis to predict adverse cutaneous drug reactions


