25 Jun Functional proteome profiling of single cells using a microchip assay
Technological advancements in sequencing technologies, as well as high-throughput methods such as droplet microfluidics, have led to more widespread use of transcriptomics in single-cell research. However, not all single-cell omics are equally advanced. For instance, although proteins can be of utmost importance in disease diagnostics, single-cell proteomics is not yet well developed. For this week’s research highlight, we have selected a recent article published in Nature Communications aiming at improving the proteomics landscape. Using microfabrication methods and a PDMS-based microchip, the research team developed a single-cell cyclic multiplexed in situ tagging (CycMIST) platform and could successfully detect 182 proteins.
“The high protein content is achieved by the multi-round labeling and multi-cycle decoding process based on the CycMIST platform. Each single cell in a microchip microwell can be stained for 4 rounds by a cocktail of UV-cleavable DNA barcoded antibodies, and for each staining the DNA oligos can be released by UV irradiation and captured by a MIST array.“, the authors explained.
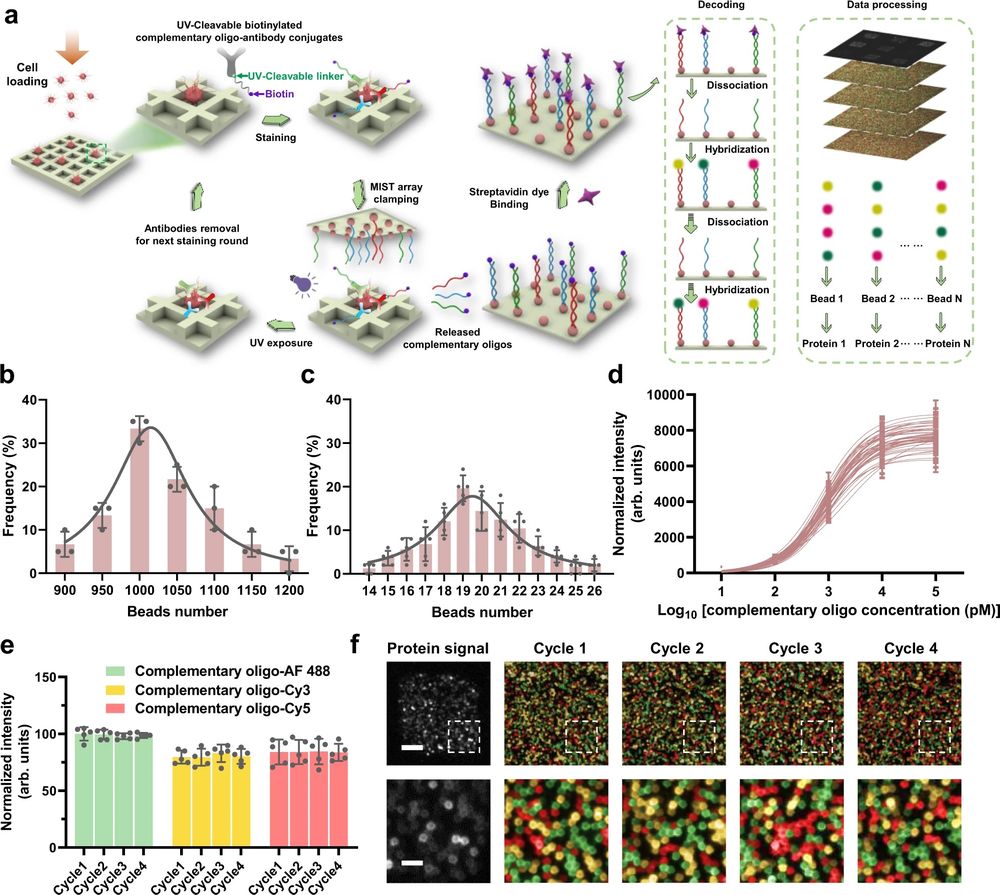
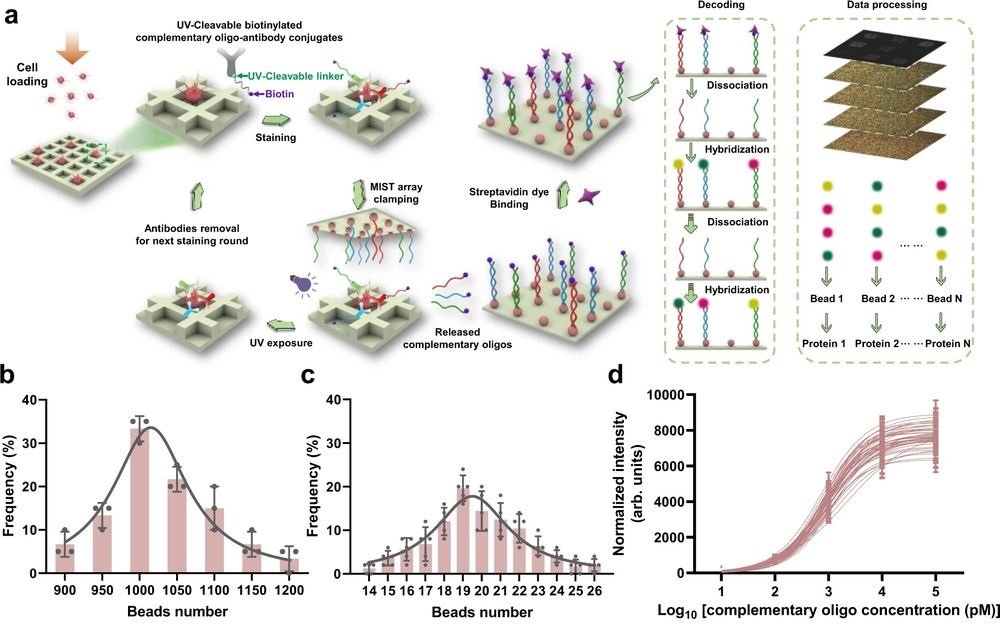
a Schematic illustration of the CycMIST process to analyze multiple proteins through the MIST microbeads array. b Distribution of the number of oligo DNA-coated microbeads on each 75 μm × 75 μm area of a MIST array that is corresponding to a PDMS microwell, n = 3 independent MIST array. c Distribution of the number of same kind oligo DNA-coated microbeads on the same MIST array, n = 5 independent MIST array. d Characterization of the CycMIST sensitivity by varying the concentrations of 50 biotinylated complementary oligo DNAs on the MIST array, n = 10 independent experiments. This is the same procedure in single-cell protein detection experiments except cell loading and conjugate binding. e Consistency of fluorescence intensities for 4 decoding cycles and for 3 fluorescent color dyes (Alexa Fluor 488, Cy3 and Cy5), n = 5 independent experiments. f Sample images of multiplexed assay of 50 proteins from a single cell by CycMIST and the 4 decoding cycles images. The greyscale images are protein detection result, and the color images are the decoding cycles from cycle 1 to cycle 4. The bottom panel is the zoom-in images from the squares in the up panel. Scale bar: 20 μm (up panel); 5 μm (bottom panel). Data are presented as mean values ± SD of more than three independent experiments, and error bars are within symbol size if not shown. The term (arb. units) is abbreviated for arbitrary units. Reproduced from Yang, L., Ball, A., Liu, J. et al. Cyclic microchip assay for measurement of hundreds of functional proteins in single neurons. Nat Commun 13, 3548 (2022). under Creative Commons Attribution 4.0 International License.
“Our microchip is amenable to integration with an instrument that can handle liquids and scan the entire array. Development of advanced data acquisition will be needed to standardize the process to be like flow cytometry, which has the hardware gating system and associated software to improve data quality and lower detection noise. A higher throughput and higher multiplexity will be achievable after further development to make the CycMIST as a tool complementary to single-cell sequencing.“, the authors concluded.
Figures are reproduced were reproduced from Yang, L., Ball, A., Liu, J. et al. Cyclic microchip assay for measurement of hundreds of functional proteins in single neurons. Nat Commun 13, 3548 (2022). https://doi.org/10.1038/s41467-022-31336-x under Creative Commons Attribution 4.0 International License.
Read the original article: Cyclic microchip assay for measurement of hundreds of functional proteins in single neurons


