23 Nov Enzymatic screening of filamentous fungi using droplet microfluidics
Abstract
“Fungal cell-wall-degrading enzymes have great utility in the agricultural and food industries. These cell-wall-degrading enzymes are known to have functions that can help defend against pathogenic organisms. The existing methods used to discover these enzymes are not well adapted to fungi culture and morphology, which prevents the proper evaluation of these enzymes. We report the first droplet-based microfluidic method capable of long-term incubation and low-voltage conditions to sort filamentous fungi inside nanoliter-sized droplets. The new method was characterized and validated in solid-phase media based on colloidal chitin such that the incubation of single spores in droplets was possible over multiple days (2–4 days) and could be sorted without droplet breakage. With long-term culture, we examined the activity of cell-wall-degrading enzymes produced by fungi during solid-state droplet fermentation using three highly sensitive fluorescein-based substrates. We also used the low-voltage droplet sorter to select clones with highly active cell-wall-degrading enzymes, such as chitinases, β-glucanases, and β-N-acetylgalactosaminidases, from a filamentous fungi droplet library that had been incubated for >4 days. The new system is portable, affordable for any laboratory, and user-friendly compared to classical droplet-based microfluidic systems. We propose that this system will be useful for the growing number of scientists interested in fungal microbiology who are seeking high-throughput methods to incubate and sort a large library of fungal cells.”

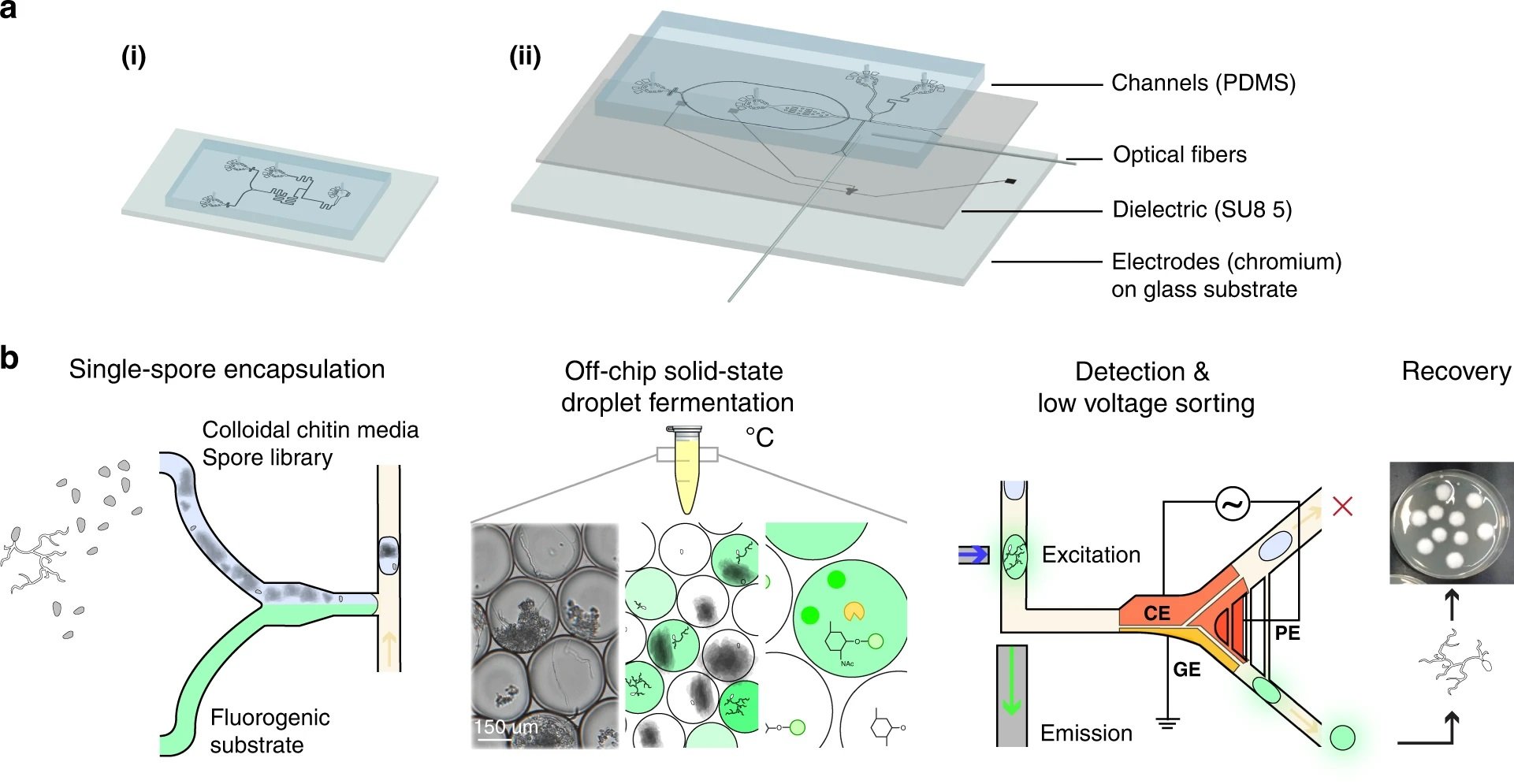
“a Schematic of two microfluidic devices used for screening: (i) a droplet generator and (ii) a sorting device containing electrodes. In (i) and (ii), the microfluidic channel is fabricated via soft lithography techniques except that (ii) contains an electrode layer with an SU-8 dielectric layer that is bonded to the channel PDMS layer. Two optical fibers (excitation: 105 μm core, 0.22 NA; emission: 200 μm core, 0.39 NA) are inserted into (ii) and placed orthogonally to the channel to excite and detect droplet fluorescence. b Screening workflow overview. Enzyme screening followed five steps. First, the mutant fungal population is generated through UV mutagenesis. Next, a microfluidic mixer T-junction droplet generator (see device (i)) co-encapsulates the conidial library suspended in colloidal chitin minimal media with a fluorescein-linked enzymatic substrate for glycoside hydrolases. The droplets were then collected and incubated in HFE7500 oil 2% fluorosurfactant at 27 °C for 2–4 days. The droplets contain single spores, minimal media with colloidal chitin, and a cell-wall-degrading specific fluorogenic substrate (FL-GlcNAc, FL-GalNAc, or FD-Glc). During incubation, glycoside hydrolase (GH) activity leads to the cleavage of the fluorescein-based substrate and the release of fluorescent fluorescein units that remain confined within the droplet. The droplet library is reinjected into the microfluidic low-voltage sorter (see device (ii)), which uses a three-electrode system (CE – constant electrode, PE – pulsing electrode, and GE – ground electrode). Mutant populations are sorted when they display high fluorescence intensity and are recovered on PDA plates and grown into clonal colonies, after which enzymatic assays are performed.” Reproduced under a Creative Commons Attribution 4.0 International License from Samlali, K., Alves, C.L., Jezernik, M. et al. Droplet digital microfluidic system for screening filamentous fungi based on enzymatic activity. Microsyst Nanoeng 8, 123 (2022).
Figures and the abstract are reproduced from Samlali, K., Alves, C.L., Jezernik, M. et al. Droplet digital microfluidic system for screening filamentous fungi based on enzymatic activity. Microsyst Nanoeng 8, 123 (2022). https://doi.org/10.1038/s41378-022-00456-1 under a Creative Commons Attribution 4.0 International License.
Read the original article: Droplet digital microfluidic system for screening filamentous fungi based on enzymatic activity


