17 Dec Enhanced Screening of Proteolytic Microorganisms Using a Passive Droplet Microfluidic Platform
Screening for microbial proteolytic activity is essential in various biotechnological applications, including bioenergy, food processing, and pharmaceuticals. Traditional methods, such as cultivation on agar plates with skimmed milk, often result in low throughput and potential false positives from non-proteolytic activities. The need for more accurate, faster, and higher-throughput methods is critical in enhancing the efficiency of microbial isolation and characterization.
“In this study, we thoroughly explored the utilization of a droplet-based system within the field of microbiology with a specific focus on the selection of proteolytic microbial strains. Key aspects of our research include: i. the design of a passive microfluidic droplet sorter, ii. the utilization of gelatin droplets for the clonal cultivation of microbes, and iii. the establishment of a novel protocol for single-cell studies and microbial isolation.“, the authors explained.
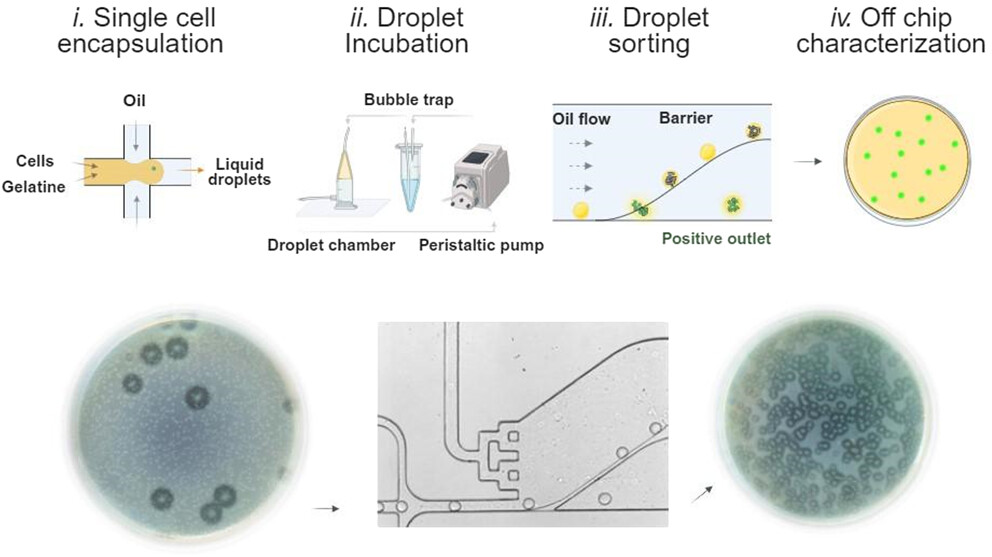
To address these challenges, the research team developed a passive droplet microfluidic platform that utilizes gelatin microgels for the encapsulation and cultivation of single microbial cells. This microfluidic device, called deformability-based passive droplet sorter (DPDS), sorts microcultures based on the deformability of droplets, which is altered by the proteolytic activity of the microbes. The microfluidic chip design allows for rapid, high-throughput screening without the need for complex instrumentation, thereby simplifying the process and reducing costs.
The microfluidic device operates by first encapsulating cells in droplets of a gelatin-based medium using a flow-focusing microfluidic droplet generator. After encapsulation, droplets are incubated to allow microbial growth, during which the proteolytic activity of the cells degrades the gelatin matrix, altering the droplet’s deformability. The microfluidic setup includes a novel passive sorting system that relies on a barrier within the chip to differentiate between more deformable droplets (indicative of proteolytic activity) and less deformable ones. This design enables a high throughput of up to 50 droplets per second, a significant improvement over previous systems.
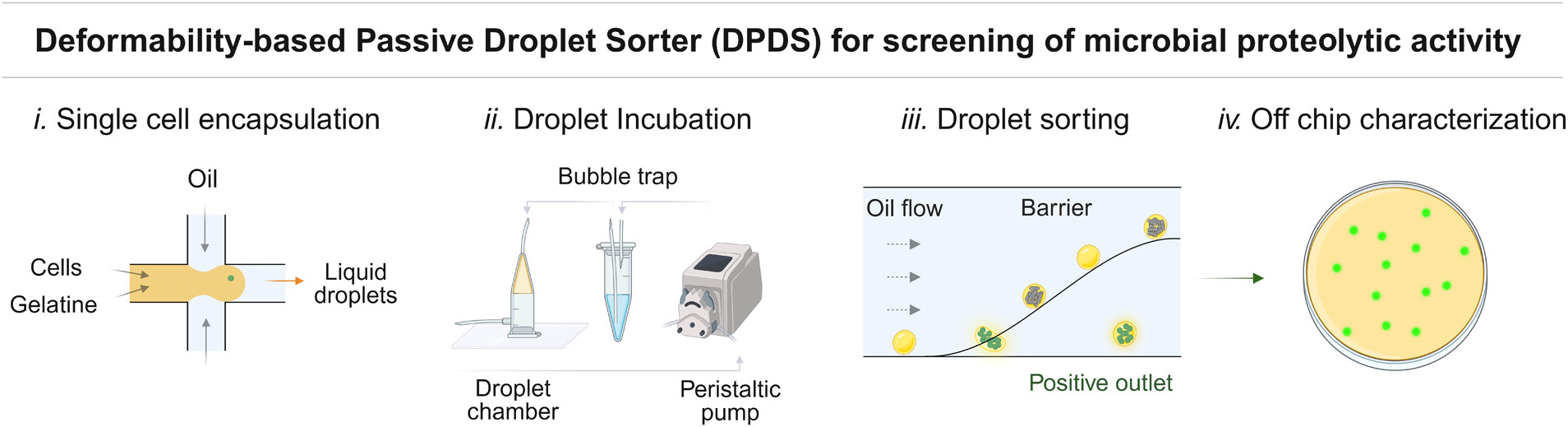
“Schematic of the high-throughput passive screening of proteolytic microbial activity. The microfluidic assay is divided into four consecutive steps: i. single bacterial cell encapsulation into liquid gelatin droplets, ii. droplet incubation in dynamic conditions, iii. sorting of droplets based on their deformability properties, iv. off-chip characterization of sorted colonies using standard microbiology methods.” Reproduced from Luca Potenza, Lukasz Kozon, Lukasz Drewniak, and Tomasz S. Kaminski Analytical Chemistry 2024 96 (40), 15931-15940. under a CC BY 4.0 Attribution 4.0 International license
The results demonstrate that the new microfluidic platform can effectively isolate proteolytic microbes with high accuracy. The method achieved an enrichment factor significantly higher than traditional methods, with minimal false positives and negatives. This increase in accuracy and throughput could greatly enhance the screening and study of microbial communities, particularly those involved in proteolysis.
This study presents a significant methodological advancement in microfluidic platforms for microbial research. By simplifying the sorting process and increasing throughput, the passive droplet-based method offers a robust tool for rapidly screening and isolating proteolytic microorganisms. This approach not only enhances the efficiency of microbial isolation but also provides a scalable solution that could be adapted for a wide range of applications in microbial screening and beyond. This work underscores the potential of integrating simple physical principles into microfluidic designs to address complex biological questions.
“Our droplet microfluidic protocol, designed for isolating proteolytic microbes, may have broader implications beyond its initial scope. The DPDS system can be used for screening hydrogels to evaluate both their polymerization and degradation processes when exposed to microbial cells. This system is not only rapid but also cost-effective, making it an ideal solution for advancing other studies, such as screening of mutant libraries of B. subtilis – a common industrial producer of proteases, or for the directed evolution of enzymes, as well as detecting the secretion of proteases by single mammalian cells. Taken together, we believe that the DPDS system could significantly advance both research and practical applications within environmental microbiology.“, the authors concluded.
Figures are reproduced from Luca Potenza, Lukasz Kozon, Lukasz Drewniak, and Tomasz S. Kaminski Analytical Chemistry 2024 96 (40), 15931-15940 DOI: 10.1021/acs.analchem.4c02979 under a CC BY 4.0 Attribution 4.0 International license.
Read the original article: Passive Droplet Microfluidic Platform for High-Throughput Screening of Microbial Proteolytic Activity
For more insights into the world of microfluidics and its burgeoning applications in biomedical research, stay tuned to our blog and explore the limitless possibilities that this technology unfolds. If you need high quality microfluidics chip for your experiments, do not hesitate to contact us.


