09 Jul DNA-programmed proteinosomes: One step further in bottom-up synthetic biology
Biological systems are regulated through networks of highly complex behaviors. Bottom-up synthetic biology provides a means for deconvoluting the underlying architecture governing these network-based behaviours. Droplet microfluidics has been the gold standard for creating complex synthetic cells from the bottom-up. To better design such systems, it is crucial to further our understanding of reaction dynamics within the compartments. For this week’s research highligh, we have selected a recent article published in Nature Communications that employed a PDMS-based microfluidic platform to study how compartmentalization and communication affect reaction rates in synthetic cell networks.
“Herein, based on our design principle, we demonstrate spatially localised PEN DNA reactions within proteinosomes. Microfluidics was used to generate mondisperse proteinosomes encapsulating a DNA template that could support designer PEN DNA reactions. In addition, this platform was used to characterise the reaction kinetics of the autocatalytic PEN DNA reaction. Due to compartmentalisation, we observe kinetic behaviours that are not accessible in buffer solutions.“, the authors explained.
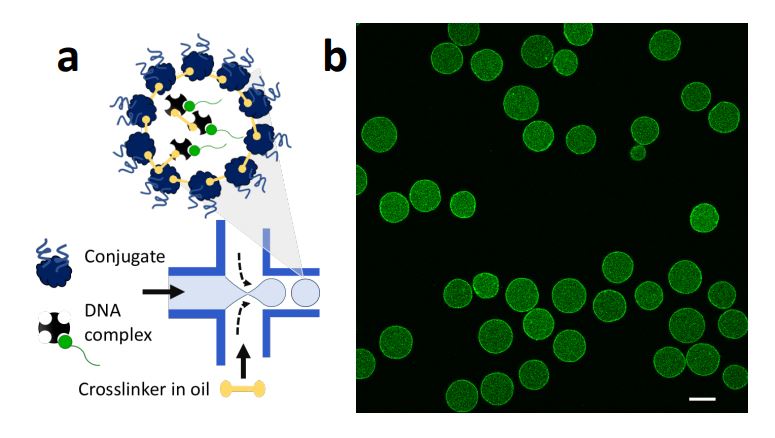
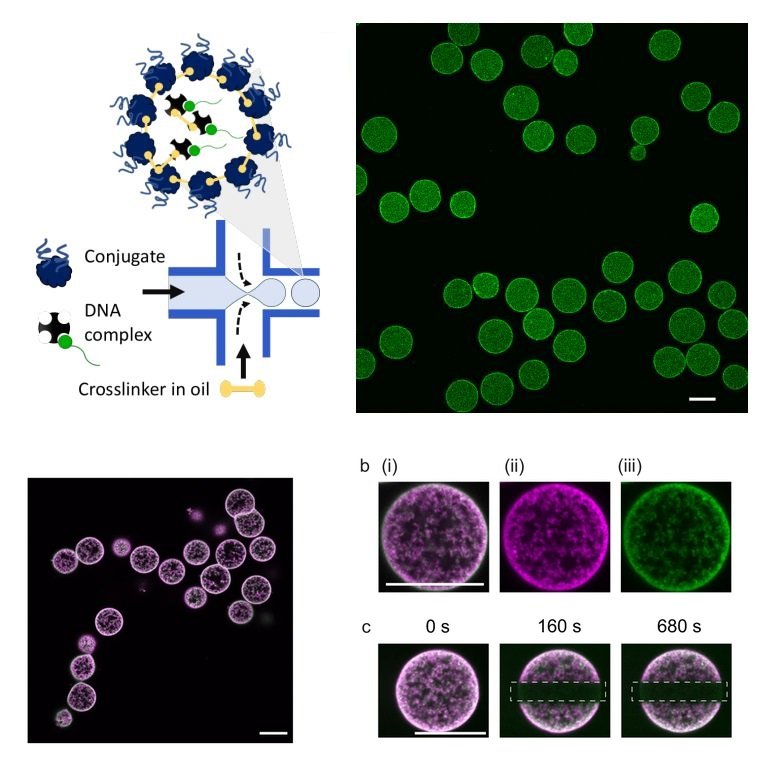
High throughput production of proteinosomes (a) A schematic illustration of the flow focusing device used for the formation of proteinosomes. Water-in-oil emulsions are produced at the flow focus junction by flowing an aqueous solution of protein conjugate (4mg.mL-1) and DNA complex (1 µM) with 2-ethyl1-hexanol containing the crosslinker BS(PEG)9 (0.5–2 mM). (b) Confocal imaging of proteinosomes containing DNA complex labelled with DY530. Scale bar is 20 µm. Reproduced from Zambrano, A., Fracasso, G., Gao, M. et al. Programmable synthetic cell networks regulated by tuneable reaction rates. Nat Commun 13, 3885 (2022). under Creative Commons Attribution 4.0 International License.
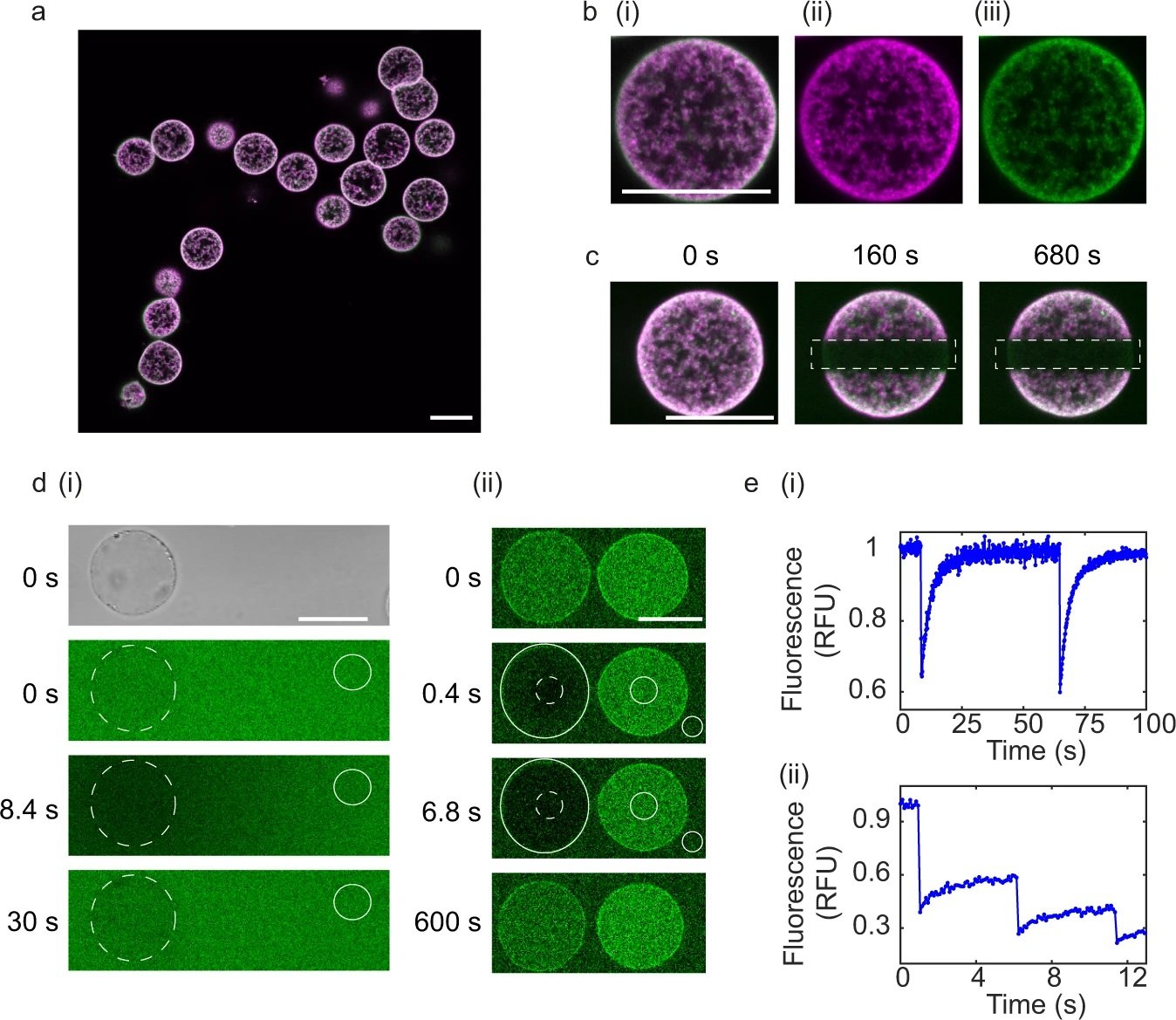
a Dual colour fluorescence confocal images of proteinosomes containing template DNA (protein conjugate labelled with fluorescein isothiocyanate (FITC) and DNA complex labelled with carboxy-X-rhodamine (ROX), b Zoom in of single proteinosome from: (i) Dual colour fluorescence, (ii) Single colour fluorescence channel, DNA labelled with ROX, (iii) protein conjugate labelled with FITC. All scale bars are 25 μm. c Fluorescent confocal microscopy images of photobleaching experiments, output frames for dual fluorescence confocal images of DNA-biotin-streptavidin complex labelled with ROX and protein conjugate labelled with FITC at t = 0 s (before bleaching), t = 160 s (after bleaching) and t = 680 s (after bleaching). Dotted rectangles show the region of photobleaching. All scale bars are 25 μm. Data are representative from at least 10 repeats. d Characterisation of the permeability of DNA (i) and PEN enzymes (ii) into proteinosomes. d (i) Bright field image (t = 0 s) used to locate proteinosomes at t = 0 s. Output frames from Fluorescence Recovery after Photobleaching (FRAP) experiments of (i) single-stranded DNA (labelled with DY530) at t = 0 s (before photobleaching), t = 8.4 s (immediately after photobleaching) and t = 30 s (after recovery). Representation from two repeats. Scale bars are 20 μm. (ii) Polymerase enzyme tagged with FITC. Output frames at t = 0 s (before photobleaching), t = 0.4 s and t = 6.8 s and t = 600 s (after recovery). Scale bar is 20 μm. Dotted circle indicates bleached spot and the non-dotted circles show backgrounds used for normalisation from a single experiment. e FRAP analysis of (i) single-stranded DNA (ii) polymerase. Source data are provided in the source data file. Reproduced from Zambrano, A., Fracasso, G., Gao, M. et al. Programmable synthetic cell networks regulated by tuneable reaction rates. Nat Commun 13, 3885 (2022). under Creative Commons Attribution 4.0 International License.
“Whilst it is still a challenge to probe the effect of compartmentalisation in biological cellular systems, the ability to build micron-sized compartments encapsulating enzyme reactions has offered an unique opportunity to address this challenge without biological complexity. Our work represents an important step in bottom-up synthetic biology approaches by combining it with quantitative approaches.“, the authors concluded.
Figures and the abstract were reproduced from Zambrano, A., Fracasso, G., Gao, M. et al. Programmable synthetic cell networks regulated by tuneable reaction rates. Nat Commun 13, 3885 (2022). https://doi.org/10.1038/s41467-022-31471-5 under Creative Commons Attribution 4.0 International License.
Read the original article: Programmable synthetic cell networks regulated by tuneable reaction rates


