16 Jan CRISPR/Cas13a incorporated into microfluidic technology for nucleic acid amplification‐free miRNA diagnostics
Although CRISPR technology is well-known for its ability for gene knock-out and therapeutic applications, it has been recently evolved as a powerful technique for diagnostics purposes. The strength of CRISPR diagnostics methods along with the superiority of microfluidics in manufacturing point-of-care (POC) diagnostics chips creates an exciting window of opportunity for developing faster and more reliable POC devices.
Recently, a research team at the University of Freiburg reported a CRISPR/Cas13a‐Powered microfluidic device for nucleic acid-amplification free miRNA diagnostics published in Advanced Materials journal. In their study, the research team led by Dr. Can Dincer and Prof. Wilfried Weber, employed the so-called “gene scissors” to identify potential brain tumour markers such as miR-19b and miR-20a from patient serum samples and in a nucleic acid-amplification free manner. To confirm the validity of the assay, they compared the results with qRT-PCR which showed a good agreement. They could achieve a readout time of 9 minutes and a total processing time of 4 hours. The limit of detection was reported to be 10pM using a measuring volume of <0.6µL.
“Our electrochemical biosensor is five to ten times more sensitive than other applications which use CRISPR/Cas for RNA analysis”, Dr. Can Dincer research group leader at the Freiburg Center for Interactive Materials and Bioinspired Technologies (FIT).
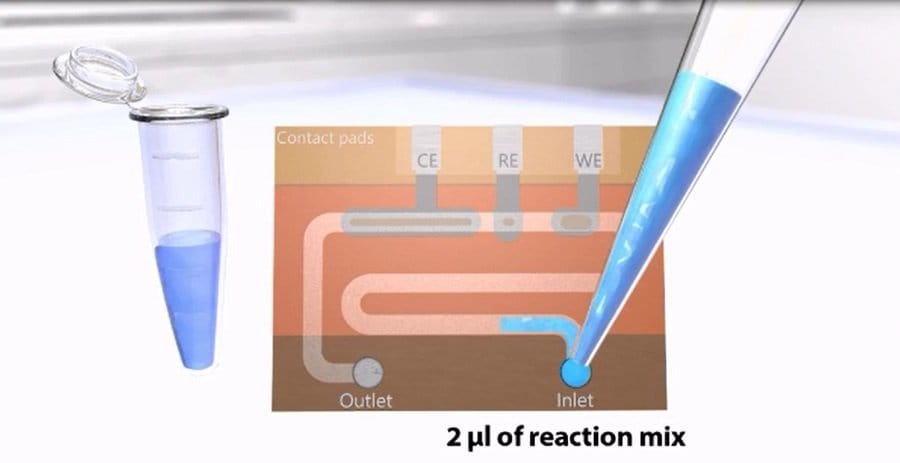
Their chip consisted of two main sections: an immobilization area and a detection region. The immobilization area was prefilled and functionalized with the required reagents. Upon incubation with the reagents, the sample is moved to the detection region that consists of three electrodes (working, counter, and reference electrodes) for electrochemical detection. The electrodes could measure the changes in the current due to the presence of H2O2 as a product of the enzymatic activity of the analytes. The amperometric peak was related to the concentration of the target miRNA, thus worked as a biosensor for measuring the miRNA content of the sample. The average cost of the chips was estimated to be <2 €. This low-cost makes this biosensor an ideal choice for third-world and developing countries with limited resources.
For future work, they have aimed at expanding the microfluidic chip to a point of care device for multiplex biosensing. The envisioned device will incorporate up to eight microchannels to allow multiple miRNA assays of the sample in a single-chip. This will have multiple benefits such as higher accuracy of the results and the possibility of performing negative and positive tests along with giving a more comprehensive picture of the patient’s condition.
This high-resolution version of the “On-chip CRISPR/Cas13a cleavage” video visualizes the “one-for-all” sensor chip handling and cleavage process and is the Supplementary video of article “CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics”, published in Advanced Materials (DOI: 10.1002/adma.201905311).
Read the original research article: CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics

Pouriya Bayat
Pouriya is a microfluidic production engineer at uFluidix. He received his B.Sc. and M.A.Sc. both in Mechanical Engineering from Isfahan University of Technology and York University, respectively. During his master's studies, he had the chance to learn the foundations of microfluidic technology at ACUTE Lab where he focused on designing microfluidic platforms for cell washing and isolation. Upon graduation, he joined uFluidix to even further enjoy designing, manufacturing, and experimenting with microfluidic chips. In his free time, you might find him reading a psychology/philosophy/fantasy book while refilling his coffee every half an hour. Is there a must-read book in your mind, do not hesitate to hit him up with your to-read list.


