22 May Advancing Tuberculosis Therapy with Microfluidic Technology
Tuberculosis (TB) remains one of the most persistent global health threats, primarily due to the ability of Mycobacterium tuberculosis (Mtb) to develop drug resistance and persist in the host for extended periods. The slow and grueling treatment process, often marred by relapse, underscores the urgent need for innovative solutions. Traditional drug discovery methods, which focus on genetic mutations and stable population traits, fall short in addressing the dynamic phenotypic variations that enable Mtb to evade treatment. Researchers developed a multi-condition microfluidic chip to tackle this challenge. This platform allows for high-throughput testing and precise environmental control, revolutionizing the approach to TB treatment. By focusing on phenotypic variations, the platform provides a detailed understanding of how individual bacterial cells respond to different treatments.
“In this work, we develop a multi-condition microfluidic platform suitable for imaging two-dimensional growth of bacterial cells during transitions between separate environmental conditions. With this platform, we implement a dynamic single-cell screening for pheno-tuning compounds, which induce a phenotypic change and decrease cell-to-cell variation, aiming to undermine the entire bacterial population and make it more vulnerable to other drugs.“, the authors explained.
The microfluidic device consists of 32 pairs of microchambers, each supplied by independent reservoirs. This design facilitates simultaneous testing of multiple compounds, significantly speeding up the experimental process. The platform’s long-term imaging capabilities enable researchers to observe bacterial growth and phenotypic changes in real-time, providing valuable insights into the bacteria’s behavior under various treatments.
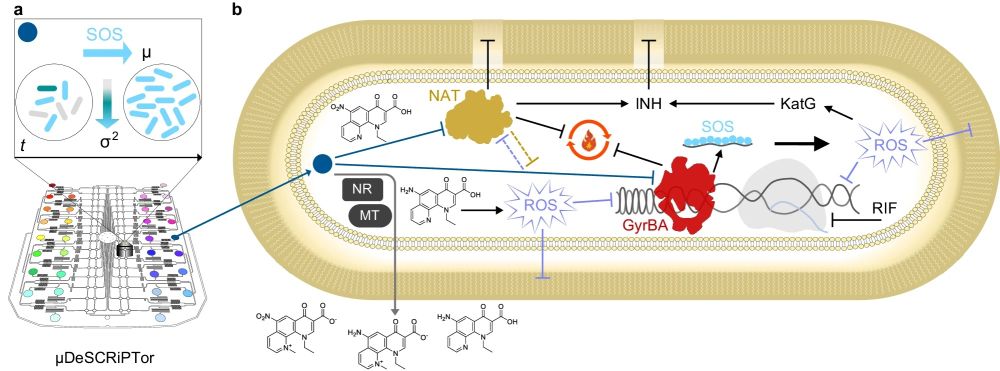
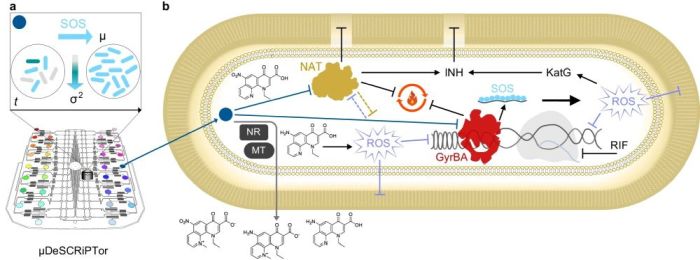
“a Schematic of the microfluidic dynamic single-cell screening for PTC (colored circles), based on induction of the SOS DNA damage response (light-blue arrow) and decrease in cell-to-cell variation (gradient arrow). The growth and phenotypic transition of rod-shaped bacteria over time (t) is also illustrated. The main PTC hit M06 is shown as a blue circle. b Schematic of the expected mode of action and drug potentiation effect of M06. Blunt arrows indicate inhibition and sharp arrows indicate induction. Blue and gray sharp arrows show the entry of M06 into the cell and the exit of its main metabolites, respectively. Bioprocessing of M06 is carried out by unknown nitroreductases (NR) and methyltransferases (MT). Nitroreduction decreases M06 potency and is a likely source of oxidative stress64. Reactive oxygen species (ROS) damage lipids, DNA, and may affect NAT (lilac flat arrows). M06 is presumed to inhibit NAT, thus affecting the composition and stability of the mycobacterial cell envelope and impairing the energy metabolism (black flat arrows). NAT might otherwise be implicated in oxidative stress tolerance (orange flat arrow). M06 inhibits DNA gyrase causing DNA breaks. The presence of single-stranded DNA triggers the SOS response. Overall, impairment of cellular energy, lipid, and DNA metabolism might induce additional oxidative stress, which further affects different macromolecules and processes in the cell. Metabolic remodeling and induction of oxidative stress response help the cell countering oxidative damage61. This includes the upregulation of the catalase peroxidase KatG, which activates the anti-tubercular drug INH32. NAT inhibition by M06 further contributes to INH activation, destabilizing the cell integrity. On the other hand, M06-mediated inhibition of negative DNA supercoiling in the presence of the anti-tubercular drug RIF, which inhibits the RNA polymerase, further impairs DNA topology, preventing both DNA replication and transcription67, with lethal consequences for the mycobacterial cell.” Reproduced from Mistretta, M., Cimino, M., Campagne, P. et al. Dynamic microfluidic single-cell screening identifies pheno-tuning compounds to potentiate tuberculosis therapy. Nat Commun 15, 4175 (2024). under a CC BY 4.0 Attribution 4.0 International license
The microfluidic device was constructed using polydimethylsiloxane (PDMS), a biocompatible material ideal for creating the intricate features needed for cell culture. The microfluidic chip integrates time-resolved microscopy to capture the behavior of live cells over time, operating on gentle hydro-pneumatic trapping to facilitate the two-dimensional growth of bacterial cells under controlled environmental conditions. The platform comprises 32 pairs of microchambers, each connected to independent reservoirs, allowing researchers to inject different solutions into each chamber and test multiple compounds simultaneously without cross-contamination.
The study identified a lead compound, M06, which increases RecA expression and reduces cell-to-cell variation, making the bacterial population more uniformly susceptible to treatment. The µDeSCRiPTor strategy allowed the researchers to pinpoint four main hits out of fewer than a hundred tested compounds that effectively pushed clonal cells into a RecA-induced state. M06 was found to impair both cellular integrity and DNA, leading to increased oxidative stress in Mtb cells. This dual mechanism of action enhances the efficacy of existing anti-tubercular drugs. Detailed spatiotemporal data obtained from the microfluidic device allowed the researchers to understand the compound’s effects more thoroughly. The lead compound M06 not only undermines the mycobacterial cell but also enhances the potency of other anti-tubercular drugs. When combined with standard antibiotics, M06 significantly reduced the survival of Mtb, demonstrating a synergistic effect that holds promise for therapeutic development.
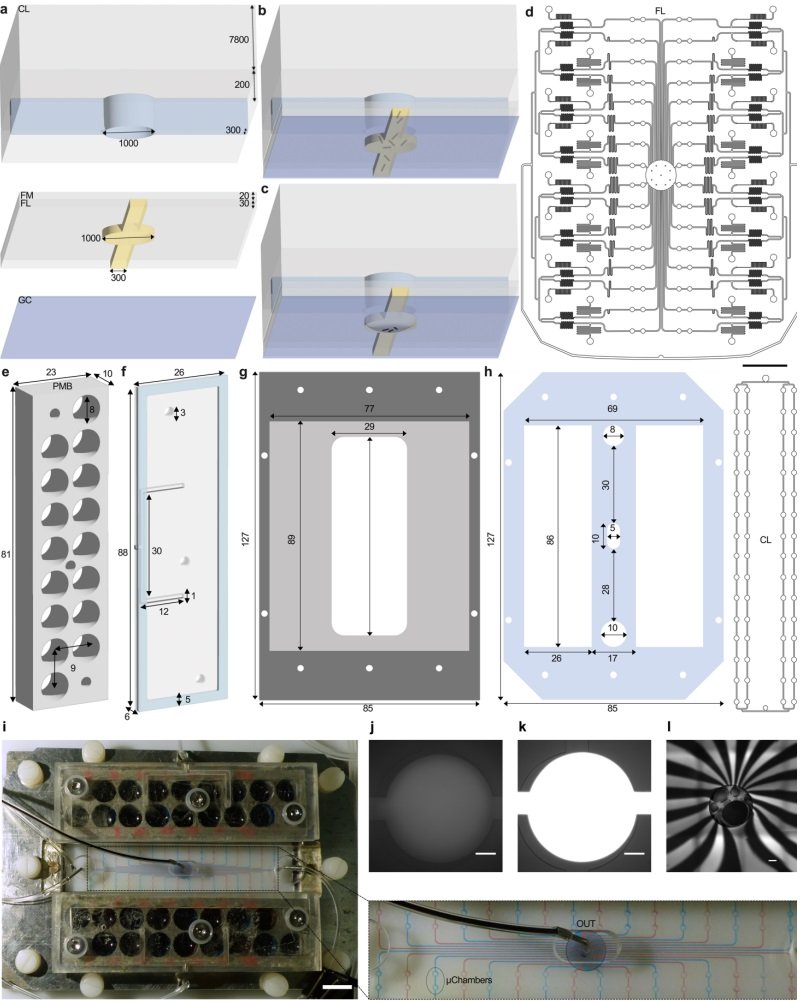
“Design and structural components of the 32-condition microfluidic platform. a−c Not-to-scale axonometric projections of the components of the microchamber (viewed from the bottom). From top to bottom (a): control layer filled with water (CL, light blue); flexible membrane (FM); flow layer filled with growth medium (FL, yellow); glass coverslip (GC). Numbers represent dimensions in µm. Microchamber assembled layers, with bacteria (black rods) perfused in the FL, without (b), or with (c) pressure applied from the CL. Bacteria are trapped between FM and GC as the pressure increases in the CL. d, Mask design of the FL and CL (Supplementary Data 1). Scale bar = 10 mm. e,f Axonometric projections of the perforated metal block (PMB) forming 32 reservoirs (e) and of its sealing lid, used for air injection (f). Numbers represent dimensions in mm. 4 g,h Schematics of the metal (g) and acrylic (h) holders used to mount the 32-condition device on the stage of an inverted wide-field microscope. The maximum area corresponds to a standard multiwell plate. Numbers indicate dimensions in mm. i, Picture of the 32-condition platform assembly, upon alternate injection of red and blue dyes from separate reservoirs. Scale bar = 10 mm. Amagnification of the core of the platform is shown (dashed rectangle), where the common outlet port (OUT, dotted circle) and a pair of microchambers (dotted oval) are also indicated. j,k,l Representative fluorescence pictures of microchambers perfused either with non-fluorescent medium (j) or with a FITC solution (k), and of the outlet port of the device (circle) receiving nonfluorescent and fluorescent medium from the outlet channels (alternate dim and bright segments) of each pair of microchambers, showing proper separation of the conditions (l). Scale bars = 200 µm” ReproducedMistretta, M., Cimino, M., Campagne, P. et al. Dynamic microfluidic single-cell screening identifies pheno-tuning compounds to potentiate tuberculosis therapy. Nat Commun 15, 4175 (2024). under a CC BY 4.0 Attribution 4.0 International license
The platform’s ability to dynamically screen for pheno-tuning compounds and monitor their effects on bacterial populations in real-time is particularly crucial for tackling pathogens that are increasingly difficult to treat. This innovative approach leverages the inherent phenotypic variation of bacterial cells, turning it into a vulnerability rather than a defense mechanism. The development and application of this microfluidic platform mark a significant milestone in TB research and treatment. It exemplifies how cutting-edge microfluidic technology can be used to address some of the most pressing challenges in global health. As we move towards a future where drug-resistant infections are a growing threat, such innovative solutions will be essential in ensuring effective and sustainable treatments.
“We expect that this approach will prove useful to identify other compounds that can enhance antimicrobial therapy, helping us to fight infections that are becoming virtually incurable.“, the authors concluded.
Figures are reproduced from Mistretta, M., Cimino, M., Campagne, P. et al. Dynamic microfluidic single-cell screening identifies pheno-tuning compounds to potentiate tuberculosis therapy. Nat Commun 15, 4175 (2024). https://doi.org/10.1038/s41467-024-48269-2 under a CC BY 4.0 Attribution 4.0 International license.
Read the original article: Dynamic microfluidic single-cell screening identifies pheno-tuning compounds to potentiate tuberculosis therapy
For more insights into the world of microfluidics and its burgeoning applications in biomedical research, stay tuned to our blog and explore the limitless possibilities that this technology unfolds. If you need high quality microfluidics chip for your experiments, do not hesitate to contact us.


