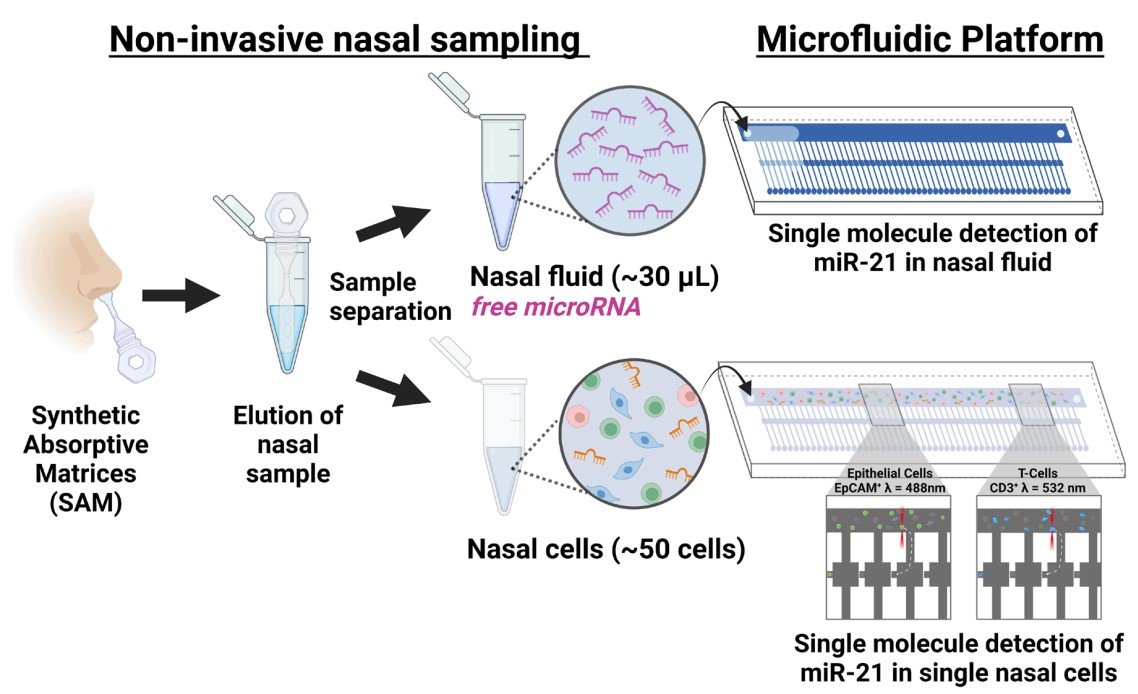
30 Apr A Complete Lab-on-a-Chip Device for Single-Cell miRNA Detection
Scientists have developed a new lab-on-a-chip device that can detect single molecules of microRNA in single cells. The device, which combines microfluidics and microarray technologies, can isolate and lyse cells and detect specific miRNA on a single chip. The method does not require nucleic acid or signal amplification, reducing experimental time and costs and potentially leading to more accurate results.
While label-free miRNA sandwich assays have been developed to quantify miRNA in human samples, they lack single miRNA molecule sensitivity in single cells. Fortunately, advancements in microfluidic single-cell analysis technology has allowed for the development of single-cell analysis methods, which can minimize the limitations of traditional bulk measurements. The new sandwich hybridisation-based assay using the miniaturised MAC chip can detect low levels of miRNA in small quantities of biological fluid, making it advantageous if cells are not available.
“Herein, we present an amplification-free sandwich hybridisation assay to detect single miRNA molecules in single cells using a microfluidic platform that optically traps and lyses individual cells. “, the authors explained.
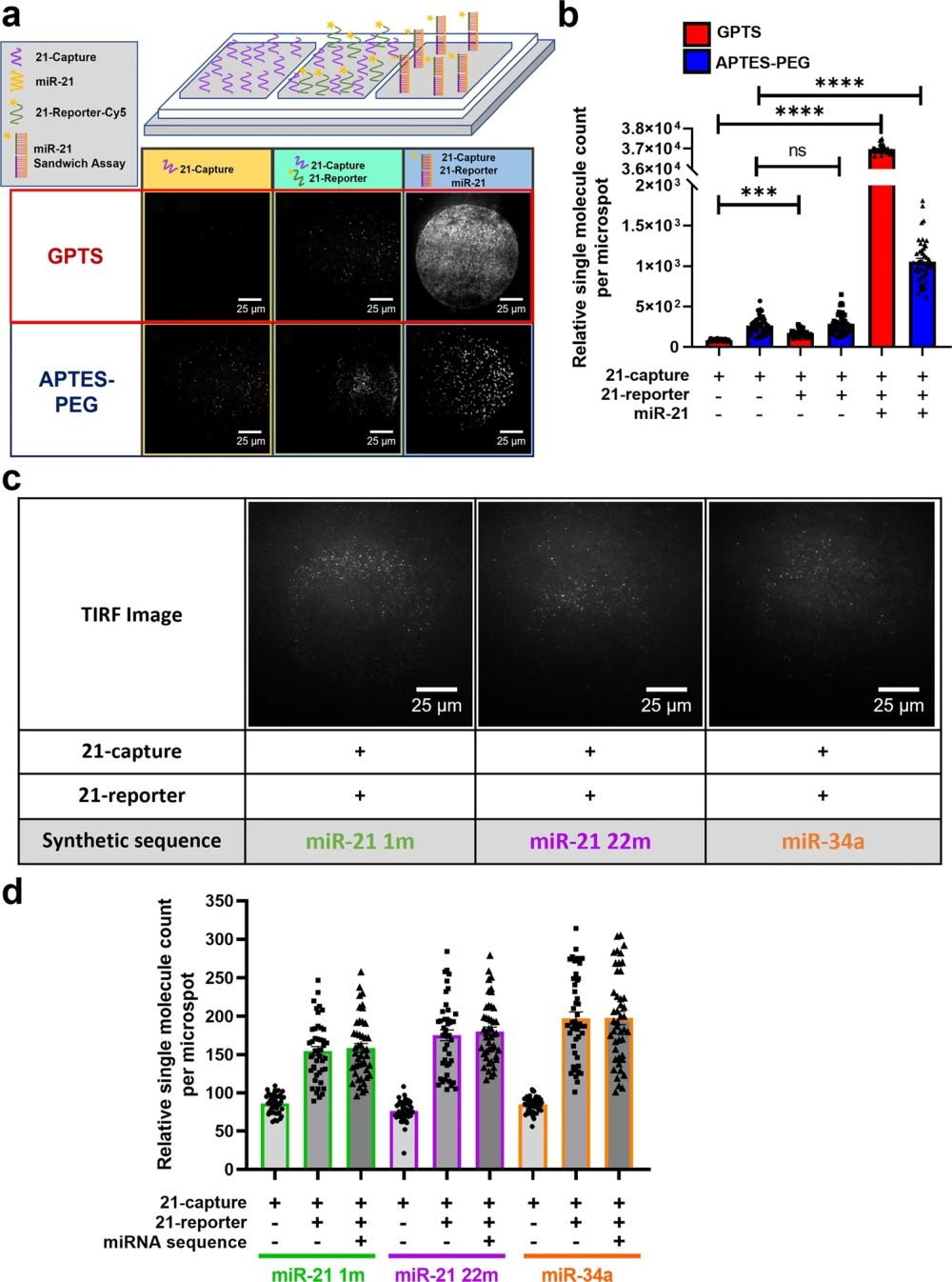
“a Schematic diagram of an open chip platform consisting of three wells comparing GPTS (Red) and APTES-PEG (Blue) passivated surfaces: first well with 21-capture probe, second well with 21-capture and 21-reporter probes, and third well containing 1.2 ×106 molecules/nL miR-21 sequences with 21-capture and 21-reporter probes. TIRF images show single molecule binding events taking place on GPTS and APTES-PEG functionalised surface within the open chip. b Bar graph of single miR-21 molecules detected using the 21-capture microarrayed spots within the open chip. Open chips were incubated and imaged every 10 min for 2 h.” Reproduced from Ho, V., Baker, J.R., Willison, K.R. et al. Single cell quantification of microRNA from small numbers of non-invasively sampled primary human cells. Commun Biol 6, 458 (2023) under Creative Commons Attribution 4.0 International License.
The microfabrication of the microfluidic chips was conducted using an SU-8-based photolithographic and PDMS soft-lithographic technique. The microfluidic chip design consists of a main microchannel connected to 55 microchannels perpendicularly that lead to individual analysis chambers. Within the microfluidic chip device, 50 chambers are microarrayed with a miRNA capture spot on the surface of the coverslip.
The microfluidic chip requires only 50 cells per experiment, compared to real-time qPCR, which typically requires ~100,000 cells/experiment. The workflow can be adapted for single-cell analysis of many biological materials, especially for studying rare cell types that are extremely precious. One can perform a multiplex assay to detect and quantify more than one biomarker simultaneously from the same cell, for example miRNA and protein expressed from a single cell to improve accuracy and diagnostic value.
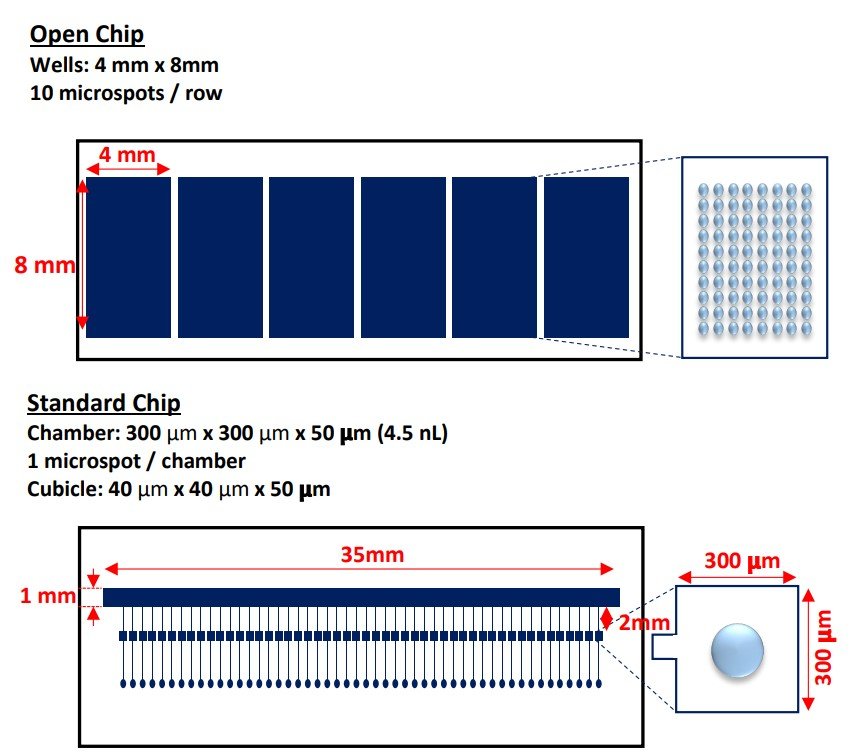
“Microfluidic chip designs.” Reproduced from Ho, V., Baker, J.R., Willison, K.R. et al. Single cell quantification of microRNA from small numbers of non-invasively sampled primary human cells. Commun Biol 6, 458 (2023) under Creative Commons Attribution 4.0 International License.
The microfluidic device could be used for detection of targets expressed in several diseases, potentially enabling a generalised single screening test. This would be a major advancement in the field of disease diagnosis and treatment, as current methods are often invasive, time-consuming, and costly. The microarray printing process is detailed in the paper and involves chemical surface passivation to minimise non-specific binding of microRNA and aid the formation of the microarray printed capture spot size. Chemical surface passivation was performed using two chemical passivation protocols: (3-Aminopropyl)triethoxysilane (APTES-PEG) and 3-Glycidyloxypropyl)trimethoxysilane (GPTS) functionalised surface. GPTS surface was found to have greater capture efficiency due to the formation of strong amide bond between the epoxide ring on the glass surface and amine group on the oligonucleotide.
In conclusion, the development of a complete lab-on-a-chip device that is sensitive to detect single molecules of miRNA in single cells via an extraction, label, and amplification-free technique from primary human samples obtained non-invasively is a significant advancement in the field of disease diagnosis and treatment. This method has the potential to be applied directly for single-cell analysis of many biological materials, especially for studying rare cell types that are extremely precious. Furthermore, the microfluidic chip device could be used for detection of targets expressed in several diseases, potentially enabling a generalised single screening test.
“This workflow can be applied directly for single-cell analysis of many biological materials, especially for studying rare cell types that are extremely precious. One can perform a multiplex assay to detect and quantify more than one biomarker simultaneously from the same cell, for example miRNA and protein expressed from a single cell to improve accuracy and diagnostic value. Furthermore, the microfluidic chip device could be used for detection of targets expressed in several diseases, potentially enable generalised single screening test.”, the authors concluded.
Figures are reproduced from Ho, V., Baker, J.R., Willison, K.R. et al. Single cell quantification of microRNA from small numbers of non-invasively sampled primary human cells. Commun Biol 6, 458 (2023). https://doi.org/10.1038/s42003-023-04845-8 under Creative Commons Attribution 4.0 International License.
Read the original article: Single cell quantification of microRNA from small numbers of non-invasively sampled primary human cells


