19 Oct Microfluidically Engineered Hydrogel Beads for Complex Protein Characterization
In the ever-evolving landscape of biochemical research, protein complexes characterization plays an important role in understanding the intricate mechanisms governing cellular functions. Traditional methods, while effective, often fall short when it comes to handling low-input samples or preserving the native states of protein complexes during analysis. However, a recent microfluidic approach has been reported in a study that introduces the Stationary-phase-dissolvable Native Affinity Purification and Mass Spectrometric characterization (SNAP-MS) technique. This method leverages biofunctionalized dissolvable hydrogel microbeads generated using droplet microfluidics to enhance the efficiency and accuracy of protein complex analysis.
“Here we introduce SNAP-MS, a Stationary-phase-dissolvable Native Affinity Purification and Mass Spectrometric characterization strategy. It allows for highly efficient purification and characterization from inputs at the pico-mole level. SNAP-MS replaces traditional elution with matrix dissolving during the recovery of captured targets, enabling the use of high-affinity bait-target pairs and eliminates interstitial voids. “, the authors explained.
SNAP-MS is centered around a novel type of biofunctionalized microbeads generated using a droplet microfluidic device, crafted from hydrogel materials that dissolve under specific conditions to release captured protein complexes in their native forms. This method elegantly circumvents the need for traditional elution processes that often disrupt protein interactions, thus preserving the structural and functional integrity of the proteins.
The process begins with the synthesis of hydrogel microbeads using a microfluidic chip containing a cross-junction, which are then chemically modified to attach bait proteins capable of capturing target protein complexes. The key innovation here is the bead’s ability to dissolve, either through chemical reactions or UV irradiation, thus freeing the bound complexes in a gentle manner that avoids disrupting their native conformation.
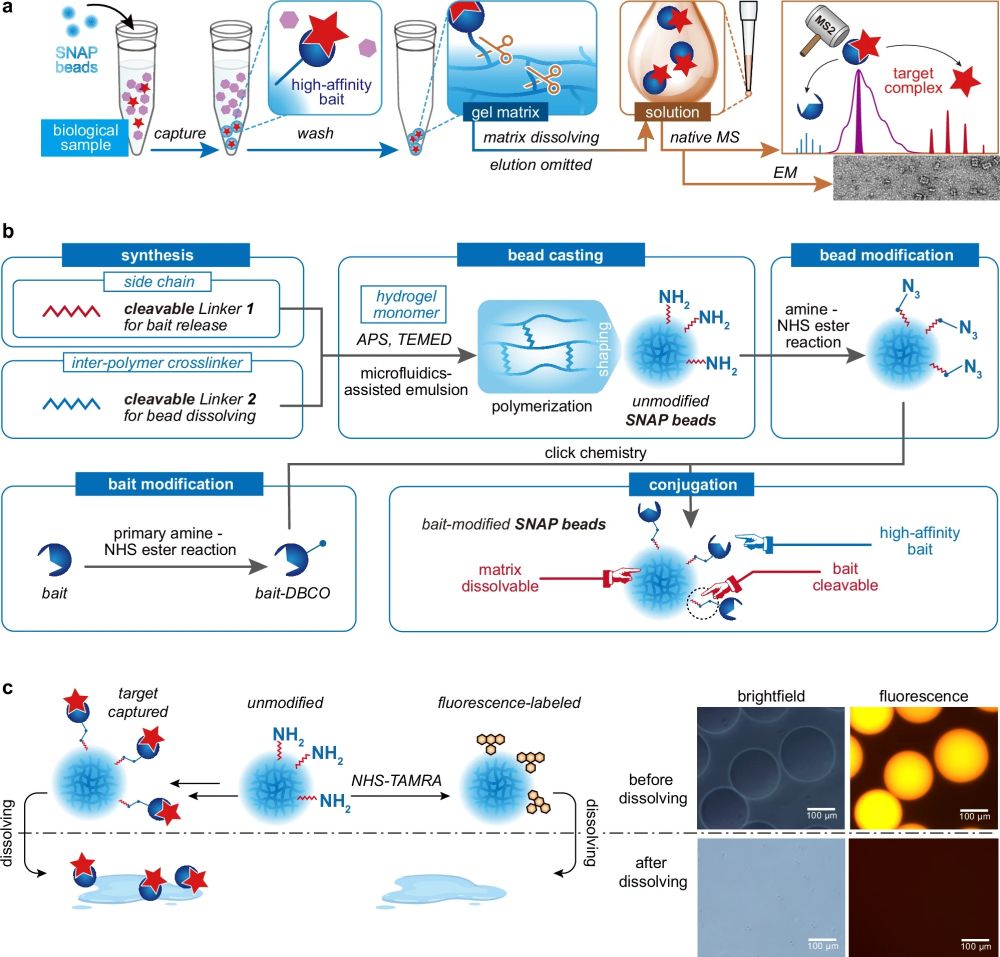
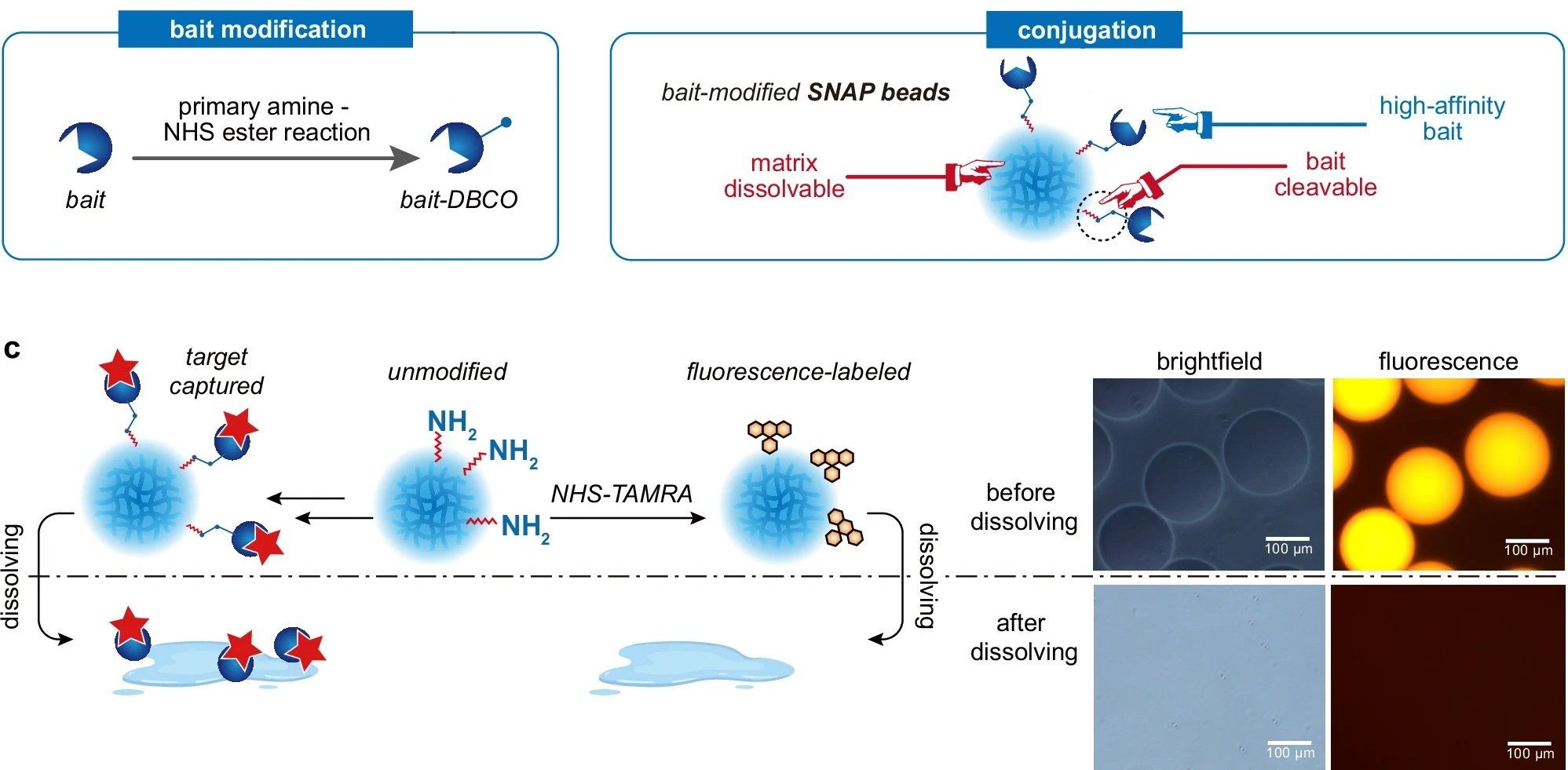
“a Protein purification and characterization workflow utilizing SNAP beads. b Fabrication procedure for SNAP beads. c Visualization of the dissolving process of fluorophore-labeled SNAP beads, characterized by both brightfield and fluorescence microscopy.a Protein purification and characterization workflow utilizing SNAP beads. b Fabrication procedure for SNAP beads. c Visualization of the dissolving process of fluorophore-labeled SNAP beads, characterized by both brightfield and fluorescence microscopy.” Reproduced from Shao, X., Tian, M., Yin, J. et al. Biofunctionalized dissolvable hydrogel microbeads enable efficient characterization of native protein complexes. Nat Commun 15, 8633 (2024) under a CC BY 4.0 Attribution 4.0 International license
Operating SNAP beads involves a series of steps designed to capture, release, and then characterize protein complexes:
Protein Capture: Proteins or protein complexes are captured from a sample by the bait-modified beads. The bait molecules, which have a high affinity for the target proteins, are previously conjugated to the beads.
Bead Dissolving: Once the proteins are bound, the bead matrix is dissolved. This can be done by exposing the beads to a specific chemical reagent that cleaves the chemical linkers or by irradiating them with UV light for photo-cleavable linkers. Dissolving the beads releases the protein complexes still bound to their bait molecules.
Mass Spectrometry Analysis: The released proteins can then be analyzed using mass spectrometry. This step is facilitated by the fact that the bait molecules attached to the proteins can act as both charge removers and mass correctors, enhancing the accuracy of the analysis.
The integration of bait attachment and matrix dissolvability in SNAP beads allows for the preservation of the native state of protein complexes, making this approach particularly effective for studying complex biological systems where maintaining interaction integrity is crucial.
Owing to the microfluidics technology in this research, one of the standout features of SNAP-MS is its ability to handle extremely low quantities of sample material—down to the pico-mole level. This is particularly advantageous for studying rare proteins or those available only in minute amounts. Additionally, the entire process from sample to data can be completed in as little as 2 hours, offering a significant improvement in time efficiency compared to traditional methods. Moreover, the microfluidic approach taken in this study is advantageous by:
Preservation of Native State: By avoiding harsh elution conditions that can disrupt protein interactions, SNAP-MS preserves the native state of protein complexes, which is often crucial for accurate functional and structural studies.
Reduction of Non-Specific Binding: The methodology reduces non-specific binding and background noise, which are common issues in mass spectrometry analyses of complex biological samples.
One of the primary proteins used in the experiments was Green Fluorescent Protein (GFP). The researchers employed GFP to demonstrate the effectiveness of SNAP beads in capturing and releasing proteins under native conditions. GFP, known for its bright green fluorescence when exposed to light in the blue to ultraviolet range, is a common reporter protein in molecular biology, making it an ideal candidate for such studies. GFP was tagged and captured using SNAP beads that had been conjugated with specific baits (like streptavidin). This setup tested the ability of SNAP beads to specifically bind and then release GFP upon dissolution of the bead matrix. Following the dissolution of the beads, the recovery rate of GFP was assessed, and its native state was analyzed using mass spectrometry to verify that its structural integrity was preserved.
The SNAP-MS technique was specifically highlighted for its capability to handle proteins with heterogeneous glycosylation. Glycosylation is a common PTM that affects protein function and stability, and the researchers demonstrated SNAP-MS’s ability to analyze these modifications accurately. While the current findings are promising, the study presents a foundational understanding of SNAP-MS’s capabilities and sets the stage for further optimizations. Future research could explore the scalability of this technique for industrial applications or its integration into existing workflows in pharmaceutical research, where understanding protein interactions is key to drug discovery and development.
“SNAP-MS demonstrates the potential to liberate structural characterization from reliance on in vitro reconstituted protein complexes, and to serve as a general strategy for characterizing natural protein complexes in studies employing biological samples.“, the authors concluded.
Figures are reproduced from Shao, X., Tian, M., Yin, J. et al. Biofunctionalized dissolvable hydrogel microbeads enable efficient characterization of native protein complexes. Nat Commun 15, 8633 (2024). https://doi.org/10.1038/s41467-024-52948-5 under a CC BY 4.0 Attribution 4.0 International license.
Read the original article: Biofunctionalized dissolvable hydrogel microbeads enable efficient characterization of native protein complexes
For more insights into the world of microfluidics and its burgeoning applications in biomedical research, stay tuned to our blog and explore the limitless possibilities that this technology unfolds. If you need high quality microfluidics chip for your experiments, do not hesitate to contact us.


