06 May High-throughput Droplet Microfluidics Enables Discovery of Novel Mini Proteins
In the continuous search to understand life’s origins and harness biotechnology’s potential, scientists have made a new discovery using high-throughput droplet microfluidics: a minimalist enzyme, derived from a library of de novo designed proteins, exhibiting remarkable biological activity. This enzyme, named mini-cAMPase, challenges our understanding of enzyme evolution and opens up new avenues for biotechnological applications.
“The ability of unevolved amino acid sequences to become biological catalysts was key to the emergence of life on Earth. However, billions of years of evolution separate complex modern enzymes from their simpler early ancestors. To probe how unevolved sequences can develop new functions, we use ultrahigh-throughput droplet microfluidics to screen for phosphoesterase activity amidst a library of more than one million sequences based on a de novo designed 4-helix bundle.“, the authors explained.
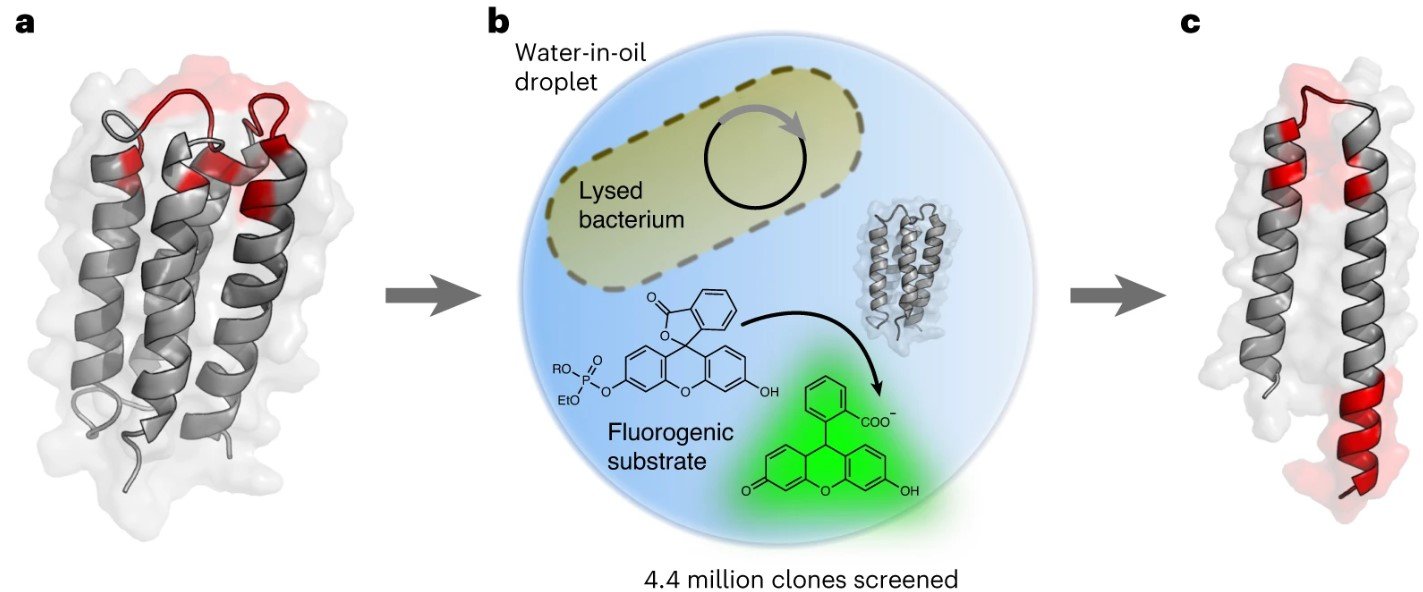
The researchers created a large library consisting of about 1.7 million variants of a de novo designed 4-helix bundle protein, termed S-824. This design allowed a high degree of sequence diversity while maintaining a basic structural framework, conducive to exploring functional possibilities. They utilized ultrahigh-throughput droplet microfluidics, a method that can test millions of variants rapidly by encapsulating each variant in tiny droplets along with substrates and reaction components. This allowed the researchers to isolate those variants that showed enzymatic activity out of the large library efficiently.

“a, De novo designed 4-helix bundle library. A library containing ~1.7 million variants based on the stably folded de novo designed 4-helix bundle protein S-824 was screened for phosphoesterase activity. The diversified residues of S-824 are shown in red (degenerate codons used: NDT/VRC/RRC). b, Ultrahigh-throughput microdroplet screening. The library was subjected to FADS on a microfluidic chip, and the 0.1–0.2% most fluorescent droplets (of a total of 4.4 million screened) were selected. c, Truncated peptide with phosphoesterase activity. The selection yielded catalytically active, truncated peptides consisting of a helix-turn-helix motif of ~60 amino acids (mutated sites are shown in red), illustrated here with a structural model of mini-cAMPase generated with AlphaFold2/ColabFold. Panel b adapted with permission from ref. 59 under a Creative Commons licence CC BY 4.0.” Reproduced from Schnettler, J.D., Wang, M.S., Gantz, M. et al. Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins. Nat. Chem. (2024) under a CC BY 4.0 Attribution 4.0 International license
Discovery Through Innovative Screening
It began with the creation of a vast library of approximately 1.7 million variants of a synthetically designed 4-helix bundle protein known as S-824. The goal was to explore how unevolved, simple protein sequences might develop into complex biological catalysts—enzymes that are fundamental to life.
Utilizing a cutting-edge technique called ultrahigh-throughput droplet microfluidics, researchers screened these millions of variants for enzymatic activity. This method involves encapsulating each protein variant in microscopic droplets along with specific substrates and chemical reagents, allowing for the rapid assessment of their catalytic properties. Through this meticulous process, one variant stood out due to its ability to efficiently hydrolyze cyclic AMP (cAMP), a crucial molecule in numerous biological signaling pathways.
Droplet microfluidic technology is a sophisticated tool that allows researchers to manipulate small volumes of fluids with high precision and control. Here’s how the microfluidic device functioned:
Droplet Generation: The microfluidic system used a flow-focusing geometry to encapsulate individual cells expressing different protein variants into droplets. These droplets, typically in the range of picoliters to nanoliters, contained a mixture of substrates and required cofactors for the enzyme reactions.
High-Throughput Screening: Each droplet acted as an independent microreactor, where the enzymatic activity could be observed and measured. The key advantage of using droplets is the ability to generate and process millions of these microreactors in a relatively short period, enabling the rapid screening of a vast library of variants.
Detection and Sorting: The microfluidic device was equipped with fluorescence detectors to identify droplets where a reaction occurred, indicated by a fluorescence signal resulting from the enzymatic activity on a fluorogenic substrate. Droplets with a higher fluorescence intensity were sorted from the rest using dielectrophoresis—a process that uses a non-uniform electric field to exert force on dielectric particles in a fluid.
Recovery and Analysis: Post sorting, the droplets containing active enzymes were collected, and the cells within them were grown for further analysis. This step was critical to confirm the activity of the enzymes and to sequence the DNA of the successful variants to identify the mutations responsible for the enzymatic activity.
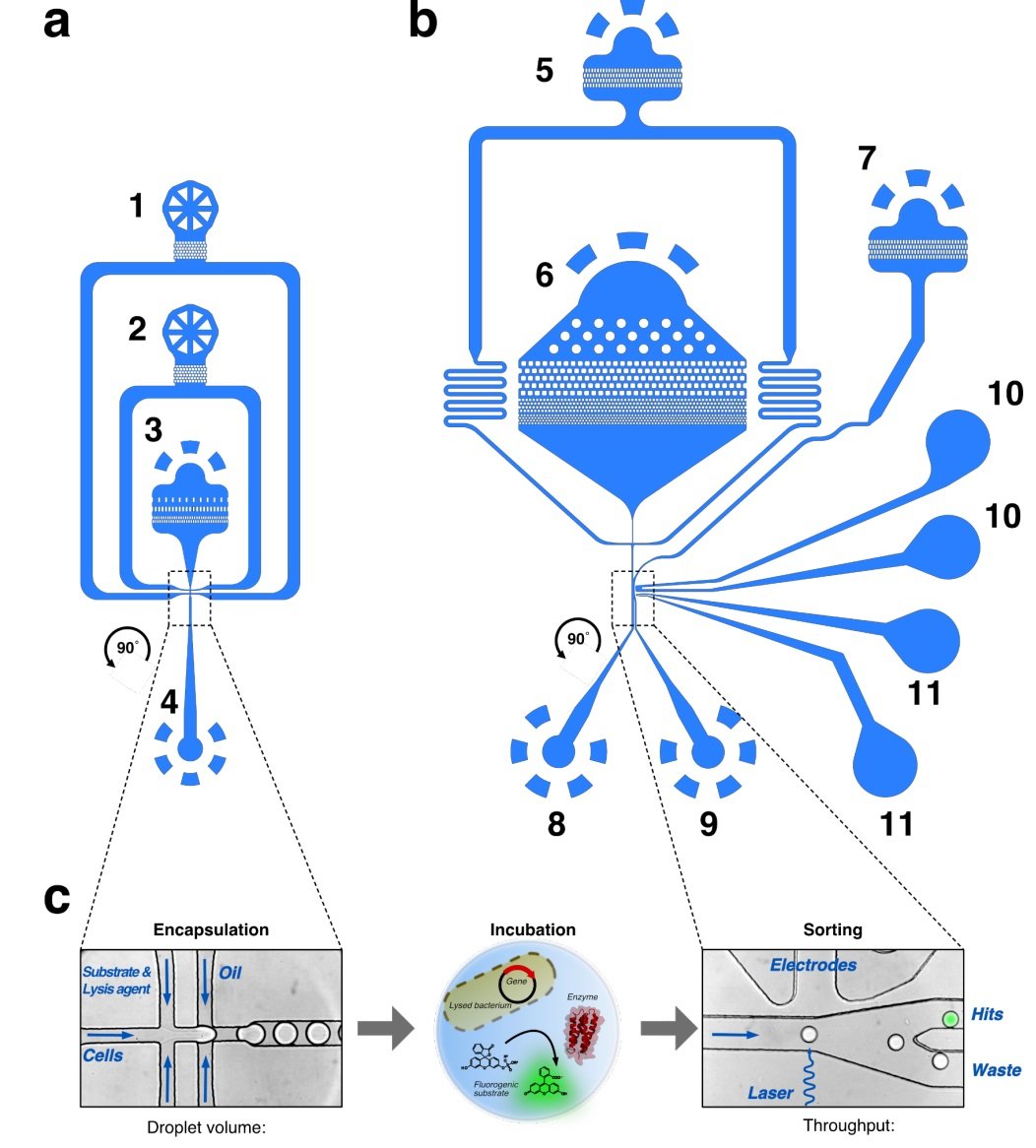
“Layouts of microfluidic chips for droplet generation and sorting. (a) Flow-focussing chip (depth: 12 µm) for droplet generation with (1) oil/surfactant mixture inlet, (2) inlet for substrate/lysis agent mixture, (3) inlet for cell suspension, and (4) outlet for droplet collection. (b) Droplet sorting chip (depth: 20 µm) for fluorescence-activated droplet sorting with (5) inlet for spacing oil, (6) inlet for droplets, (7) oil extractor, (8) waste outlet, (9) hit outlet, (10) ground electrode (+), and (11) signal electrode (–). (c) On the flow-focusing chip, E. coli cells can be co-encapsulated with a fluorogenic substrate and lysis agent into monodisperse picolitre-sized droplets. After incubation, the library-containing emulsion can be screened on the sorting chip, where the droplets pass through a sorting junction with an excitation laser and a fluorescence detector. Upon surpassing a pre-set fluorescence threshold, specific droplets can be electrophoretically sorted into the hit channel for subsequent recovery. The general layout of panels (a) and (b) of this figure is inspired by the work of Neun et al/, panel (c) is adapted from Schnettler et al. . Files of these chip designs are available for download from our repository DropBase (https://openwetware.org/wiki/DropBase) and in the Supplementary Data.” Reproduced from Schnettler, J.D., Wang, M.S., Gantz, M. et al. Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins. Nat. Chem. (2024) under a CC BY 4.0 Attribution 4.0 International license.
The identified enzyme, mini-CAMPase, exhibited remarkable traits. It was not just any enzyme; it was significantly truncated, retaining only 59 residues. Despite its reduced size, it formed a dimer—a complex of two protein molecules—that displayed a new and dynamic α-helical structure essential for its catalytic function. The enzymatic activity of mini-CAMPase was dependent on the presence of manganese ions, which is indicative of a metalloenzyme. The manganese was crucial for catalysis but did not affect the thermal stability of the enzyme, suggesting a specific catalytic role for the metal..
“Although large modern enzymes are restricted by an epistatic burden that causes mutations to interfere with structural and functional innovation76,77,78,79, small dynamic proteins could escape the limits imposed by cooperative effects and become functionally more versatile and more evolvable by reducing the cost of innovation. These patterns may reflect the role of smaller, structurally versatile peptides in Dayhoff’s model12,13. Here, functional proficiency was found by seemingly going back to the origins of life, paradoxically reaching improvements in primitive rather than sophisticated scaffolds that, as low-probability events, nevertheless become accessible via ultrahigh-throughput screening.“, the authors concluded.
Figures are reproduced from Schnettler, J.D., Wang, M.S., Gantz, M. et al. Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins. Nat. Chem. (2024). https://doi.org/10.1038/s41557-024-01490-4 under a CC BY 4.0 Attribution 4.0 International license.
Read the original article: Selection of a promiscuous minimalist cAMP phosphodiesterase from a library of de novo designed proteins
For more insights into the world of microfluidics and its burgeoning applications in biomedical research, stay tuned to our blog and explore the limitless possibilities that this technology unfolds. If you need high quality microfluidics chip for your experiments, do not hesitate to contact us.


