10 Aug Transforming Single-Cell RNA Sequencing: Microfluidic Innovations for Comprehensive Cell Analysis
Single-cell RNA sequencing (scRNA-seq) has ushered in a new era of biological understanding by enabling the assessment of gene expression profiles at the individual cell level. This advancement has illuminated the complexities of cellular heterogeneity and functional diversity within biological systems. Yet, the pursuit of enhanced efficiency, accuracy, and scalability persists. In this blog post, we delve into a remarkable marriage between scRNA-seq and microfluidic technology, a fusion that could reshape our comprehension of cellular behaviors.
“Single-cell RNA sequencing (scRNA-seq) has ushered in a new era of biological understanding by enabling the assessment of gene expression profiles at the individual cell level. This advancement has illuminated the complexities of cellular heterogeneity and functional diversity within biological systems. Yet, the pursuit of enhanced efficiency, accuracy, and scalability persists. In this blog post, we delve into a remarkable marriage between scRNA-seq and microfluidic technology, a fusion that could reshape our comprehension of cellular behaviors.”, the authors explained.
The Integration of Microfluidic Technology and scRNA-seq
A cadre of researchers has recently unveiled an ingenious approach to single-cell analysis that amalgamates the prowess of microfluidic chips with scRNA-seq methodologies. Microfluidics, with its capacity to manipulate minute volumes of fluids within microscale channels, has traversed diverse scientific domains. This study introduces the “spinDrop” rig, an innovative microfluidic platform designed to confer precision in cell encapsulation, sorting, and subsequent analysis.
A Close Examination of the spinDrop Rig
The core of this revolutionary approach is the spinDrop rig, a comprehensive system tailored for fluorescence-activated droplet sorting (FADS). Central to this system is the creation of water-in-oil droplets, each encapsulating an individual cell alongside barcoded beads. These beads host distinctive molecular barcodes that are indispensable for downstream cell identification. The rig seamlessly amalgamates fluorescence excitation and detection through optical fibers, high-speed cameras, and microfluidic devices, rendering it a holistic solution for single-cell analysis.

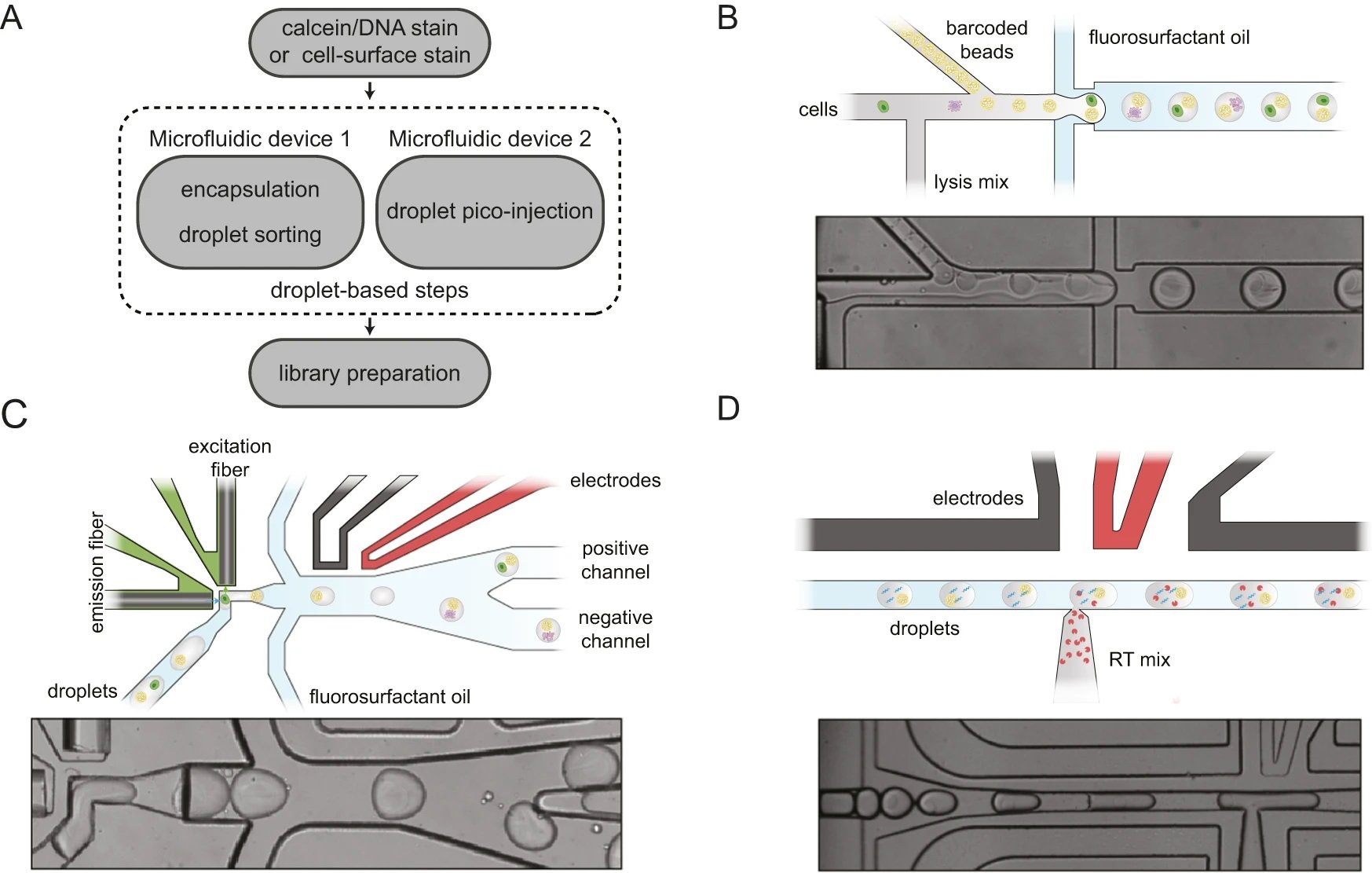
“A Schematic of the different steps to generate sequencing-ready libraries. First, intact whole cells are made detectable with a Calcein-AM viability stain, cell-surface marker stain, and intact nuclei using a DNA staining dye (such as the Vybrant DNA stain). Then the cells or nuclei are co-encapsulated with barcoded polyacrylamide beads, and droplets with viable cells, intact nuclei or specific cell types are enriched after encapsulation using in-line fluorescence-activated droplet sorting (FADS), hereby discarding empty droplets or droplets containing damaged material. The droplets containing the viable cells or intact nuclei are then lysed and heat-treated, and further re-injected in a second microfluidic device, which will inject the reverse transcriptase mix using coalescence-activated picoinjection. B Schematic of the cell/nuclei and polyacrylamide bead droplet co-encapsulation process in the first microfluidic device (top). Brightfield image of the co-encapsulation process (bottom). C Schematic of fluorescence-activated sorting of droplets containing viable cells or intact nuclei from the pool of empty droplets or droplets containing damaged or unwanted material (top). Brightfield image of the in-line sorting process (bottom). D Schematic of the coalescence-activated droplet picoinjection of a reverse transcriptase mix (top). Brightfield image of the picoinjection process (bottom).” Reproduced from De Jonghe, J., Kaminski, T.S., Morse, D.B. et al. spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content. Nat Commun 14, 4788 (2023).
The Power of Picoinjection
Picoinjection, a breakthrough technique, stands as a testament to the precision achievable in single-cell analysis. This technique involves the minute injection of reagents—enzymes, antibodies, or other bioactive molecules—directly into individual cells encapsulated within the droplets. The integration of picoinjection within the spinDrop rig equips researchers with an unprecedented ability to modify the cellular microenvironment, capture dynamic cellular responses, and unlock the intricacies of cell signaling pathways.
Critical Components of the System
1- Microscope and Laser Configuration: An inverted microscope equipped with a 488 nm laser constitutes the excitation source for fluorescence. This pivotal step enables the differentiation of various cell types based on their unique fluorescent markers.
2- Integration of Optical Fibers: The harmonization of optical fibers with the microfluidic chip is of paramount importance to ensure meticulous control over fluorescence excitation and detection. These optical fibers are meticulously prepared and integrated into the chip to optimize the efficacy of light transfer and fluorescence collection.
3- Microfluidic Chip: The microfluidic chip serves as the linchpin of the system, facilitating droplet generation, cell encapsulation, and sorting. Its design is meticulously orchestrated to accommodate optical fibers and fluidic pathways, both critical for seamless single-cell analysis.
4- Pulse Generator and High-Voltage Amplifier: For the sorting of droplets, a pulse generator and high-voltage amplifier are deployed. These components generate a brief but potent pulse that redirects droplets containing cells of interest into a distinct collection channel.
The Workflow: Encapsulation and Sorting
The workflow comprises several pivotal stages:
Droplet Generation: Leveraging microfluidic technology, water-in-oil droplets are formed. Each droplet encapsulates a singular cell along with barcoded beads. The molecular barcodes on these beads serve as unique identifiers during downstream analysis.
Fluorescence Detection: Optical fibers coupled with high-speed cameras facilitate real-time fluorescence detection emitted by cells. This permits the identification of cells based on specific fluorescent markers.
FADS Sorting: Upon fluorescence detection, a precise pulse is applied to the microfluidic device, segregating droplets containing cells of interest into a dedicated collection channel. This sorting mechanism ensures the isolation of pertinent cells for subsequent analysis.
Library Construction: Post-sorting, the isolated cells undergo RNA extraction and library construction. The molecular barcodes embedded within the barcoded beads furnish invaluable insights into the gene expression profiles of individual cells.
Technical Insights from Results
The amalgamation of microfluidic technology with scRNA-seq has reaped notable outcomes. The researchers effectively scrutinized intricate cell mixtures, capturing gene expression profiles with unparalleled sensitivity and specificity. Critically, the technology exhibited an extraordinary capability to discern rare cell types that might evade detection through conventional means.
Moreover, the research team delved into the intricacies of specific cell populations within complex tissues, unearthing concealed cellular diversities. This revelation bears profound implications for the domains of developmental biology, disease investigation, and potential therapeutic avenues.
Conclusion
The fusion of microfluidic technology and scRNA-seq represents a significant step forward in the field of single-cell analysis. The spinDrop rig exemplifies the innovative spirit of researchers striving to refine existing techniques and push the boundaries of what’s possible in biological research. As this technology continues to mature, it could reshape our understanding of cellular heterogeneity, disease progression, and therapeutic interventions.
“In summary, this work identifies a highly efficient mechanism for platelet generation outside of the body, by repeated passage of MKs through lung vasculature under air ventilation, involving enucleation and final TPM4-dependent steps to generate platelets. The findings will inform new approaches, such as the microfluidic system reported here, to large scale generation of human platelets.“, the authors explained.
Remember to check our blog for more exciting research highlights and stay up to date with the latest advancements in science and technology!
Figures are reproduced from De Jonghe, J., Kaminski, T.S., Morse, D.B. et al. spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content. Nat Commun 14, 4788 (2023). https://doi.org/10.1038/s41467-023-40322-w under a Creative Commons Attribution 4.0 International License)
Read the original article: spinDrop: a droplet microfluidic platform to maximise single-cell sequencing information content


