06 Feb Revolutionizing biomarker detection: The power of microfluidic sensing
In the field of biomolecular analysis and diagnostics, the detection of proteins plays a critical role. Typically, conventional immunosensing assays rely on surface-capture of target molecules, which can limit specificity, sensitivity, and the ability to provide information beyond simple concentration measurements. In this week’s research highlight, we delve into a recent paper published in Nature Communications that introduces a new way of directly detecting proteins in solution using a surface-free, single-molecule microfluidic sensing platform known as the digital immunosensor assay (DigitISA). The DigitISA platform combines microchip electrophoretic separation with single-molecule detection to achieve absolute quantification of proteins in a single step, providing a new experimental paradigm for protein biomarker sensing.
“we demonstrate that the assay provides information beyond stoichiometric interactions, and enables characterization of immunochemistry, binding affinity, and protein biomarker abundance. Taken together, our results suggest a experimental paradigm for the sensing of protein biomarkers, which enables analyses of targets that are challenging to address using conventional immunosensing approaches.“, the authors explained.
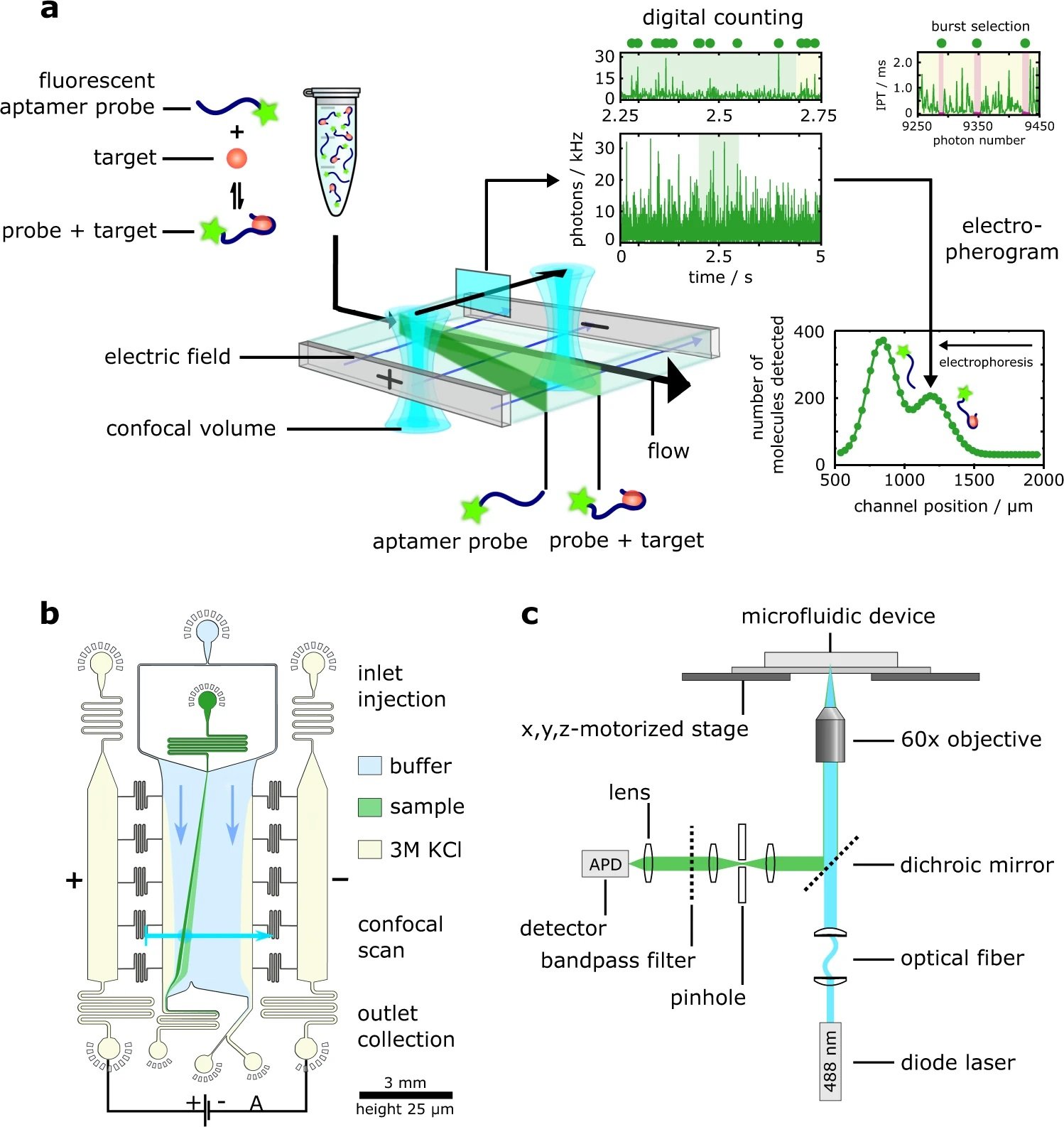
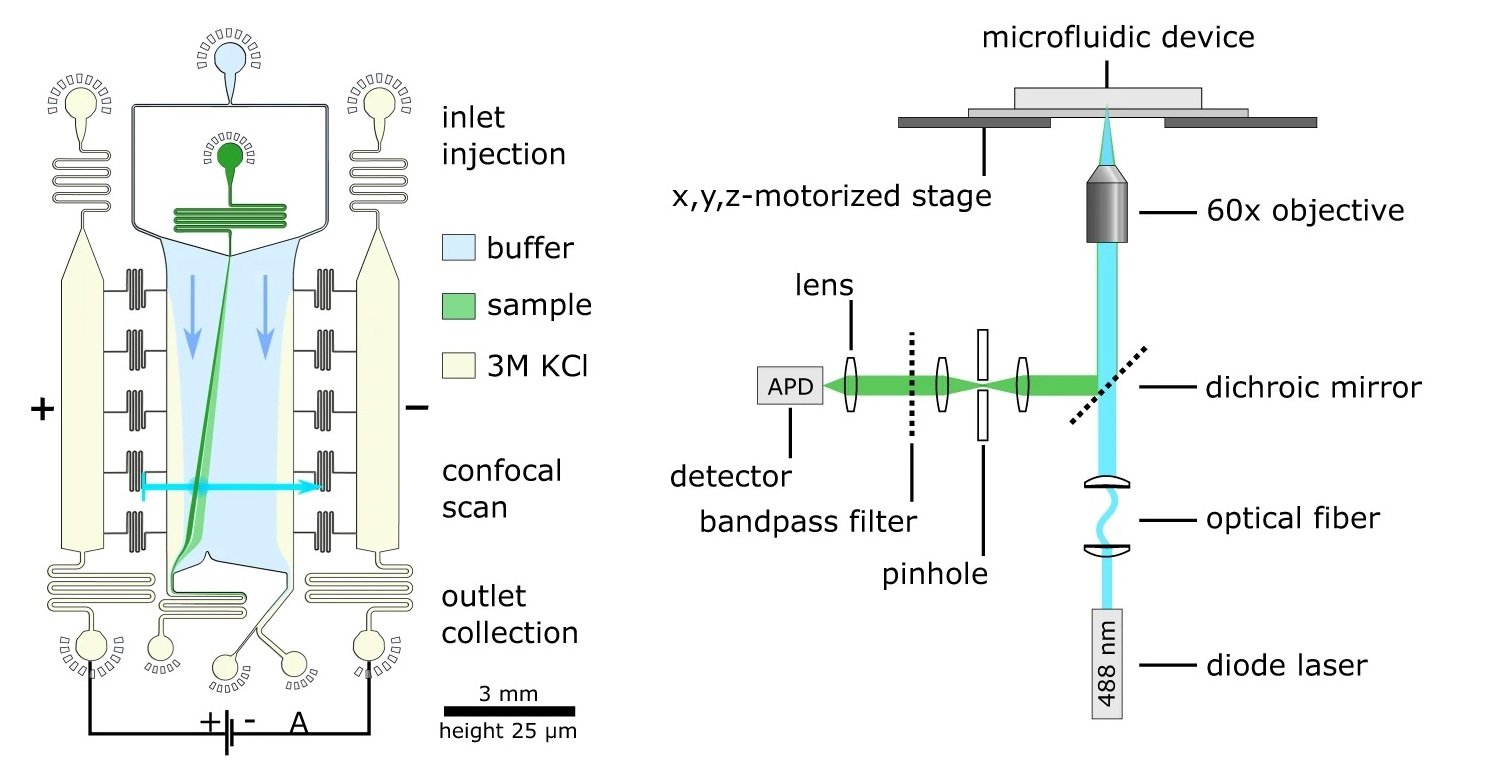
“a Schematic illustration of the DigitISA platform and overview of the experimental workflow. DigitISA integrates electrophoretic separation and single-molecule detection in a platform for single-step sensing of target proteins in solution using only a single affinity reagent. The sample including a mixture of the target protein and its fluorescently labeled affinity probe (e.g., aptamer) is injected into a micron-scale electrophoretic separation unit. The application of an electric field allows protein-bound probe molecules to be discriminated from those probe molecules that are not bound to the protein target, owing to a difference in their electrophoretic mobilities. Confocal scanning across the separation chamber is performed and the number of molecules traversing the confocal volume at each of the scanned positions is estimated ‘digitally’ from the recorded photon-count time trace using a combined inter-photon time (IPT) and photon-count threshold burst-search algorithm. From the obtained counts, an electropherogram is created allowing for a discrimination between protein-bound affinity probe and free probe. b Design of the free-flow electrophoresis device. Sample is flown into the microfluidic chip by the central injection port where it is then surrounded by the carrier buffer solution. The electrophoresis chamber is connected to a co-flowing electrolyte solution (3 M KCl) via bridges, which allows for a narrow sheet of electrolyte to flow along the sides of the chamber. An electric field is applied from metal clips at the outlets of the electrolyte channels, which propagates along the electrolyte sheet and enables separation of molecules perpendicular to the flow direction. c Schematic of the confocal microscopy setup used for single-molecule detection. A diode laser is used to excite the sample through an objective, and a single-photon counting avalanche photodiode (APD) is used to register emitted photons from the sample. The confocal volume is moved across the cross-section of the chip in a stepwise manner with the aid of a motorized stage. This allows the flux of the protein-bound probe molecules to be estimated. Details of the setup are described in the Methods section (Data analysis).” Reproduced under Creative Commons Attribution 4.0 International License from Krainer, G., Saar, K.L., Arter, W.E. et al. Direct digital sensing of protein biomarkers in solution. Nat Commun 14, 653 (2023).
DigitISA is a game-changing microfluidic device that brings a new approach to protein biomarker detection. Unlike traditional immunosensing assays, which rely on surface-capture of target molecules, the DigitISA platform is based on microchip electrophoretic separation and single-molecule detection, making it a surface-free solution for direct protein biomarker detection. This microfluidic chip provides absolute number and concentration quantification of proteins in a single step, delivering highly sensitive results in the picomolar range. The ability to perform immunochemistry analysis, determination of binding affinities, and characterization of protein biomarker abundance is what sets DigitISA apart from traditional surface-based assays. With potential for multicolor single-molecule spectroscopy and FRET techniques, as well as other microfluidic separation modalities, the DigitISA platform is poised for further development and optimization. While it currently requires expertise in advanced microfluidics and single-molecule optics, DigitISA holds promise as a commercial benchtop instrument.
The DigitISA approach releases fundamental constraints of conventional immunosensing, enabling quantitative protein biomarker sensing even at low concentrations and with low-affinity capture probes. In this work, aptamers were used as affinity probes for their higher electrophoretic mobility, recognition capabilities, and ease of production. The platform also provides direct quantitation of target concentration, characterization of immunochemistry/valency and stoichiometry, and the possibility for future development of antibody-based probes. Despite its limitations, such as the requirement for a probe and the lack of high throughput, DigitISA is a new experimental paradigm for protein biomarker sensing with broad applicability in biomolecular analysis and diagnostics. It opens the door for sensitive multimodal analysis in situations where traditional assays do not work well, paving the way for DigitISA to become an orthogonal tool in the field.
“Taken together, the microchip DigitISA platform presented herein constitutes a new experimental paradigm for protein biomarker sensing. We have demonstrated that the method is at least on par in terms of sensitivity with more traditional assay (i.e., in the picomolar range), yet with a number of qualitative advances.“, the authors explained.
Figures and the abstract are reproduced from Krainer, G., Saar, K.L., Arter, W.E. et al. Direct digital sensing of protein biomarkers in solution. Nat Commun 14, 653 (2023). https://doi.org/10.1038/s41467-023-35792-x
Read the original article: Direct digital sensing of protein biomarkers in solution


