11 Jan Detection and gene expression profiling of HIV-infected cells using droplet microfluidics
Abstract
“Rare CD4 T cells that contain HIV under antiretroviral therapy represent an important barrier to HIV cure, but the infeasibility of isolating and characterizing these cells in their natural state has led to uncertainty about whether they possess distinctive attributes that HIV cure-directed therapies might exploit. Here we address this challenge using a microfluidic technology that isolates the transcriptomes of HIV-infected cells based solely on the detection of HIV DNA. HIV-DNA+ memory CD4 T cells in the blood from people receiving antiretroviral therapy showed inhibition of six transcriptomic pathways, including death receptor signalling, necroptosis signalling and antiproliferative Gα12/13 signalling. Moreover, two groups of genes identified by network co-expression analysis were significantly associated with HIV-DNA+ cells. These genes (n = 145) accounted for just 0.81% of the measured transcriptome and included negative regulators of HIV transcription that were higher in HIV-DNA+ cells, positive regulators of HIV transcription that were lower in HIV-DNA+ cells, and other genes involved in RNA processing, negative regulation of mRNA translation, and regulation of cell state and fate. These findings reveal that HIV-infected memory CD4 T cells under antiretroviral therapy are a distinctive population with host gene expression patterns that favour HIV silencing, cell survival and cell proliferation, with important implications for the development of HIV cure strategies.”

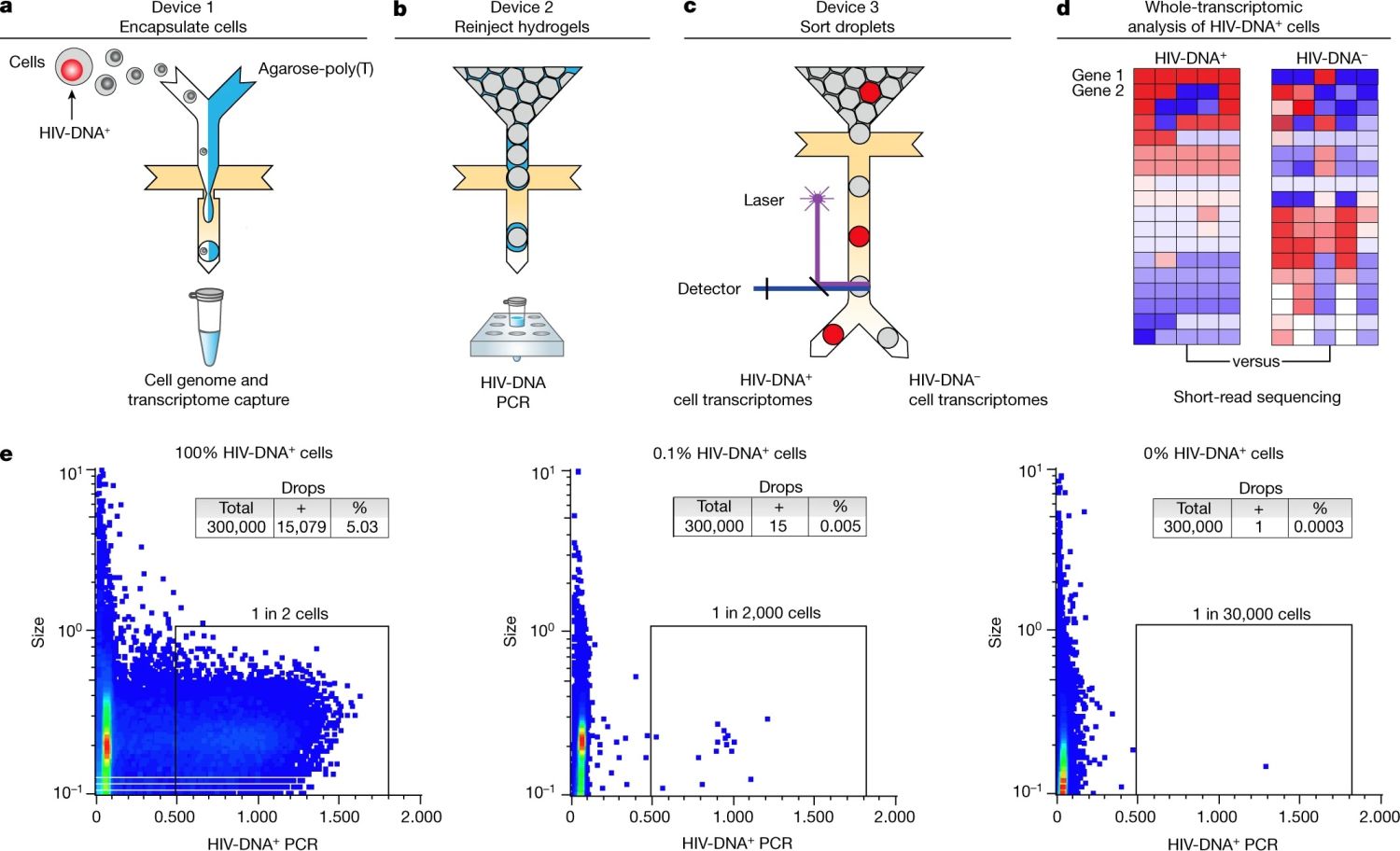
“a–d, Overview of the workflow, including three sequential microfluidic devices separated by handling steps. a, On the first device, single cells are encapsulated at a limiting dilution in a water-in-oil emulsion with lysis buffer and molten agarose-poly(T). The agarose is then cooled to form a hydrogel bead that retains genomic DNA and polyadenylated RNA. After oil removal, whole-transcriptome cDNA is covalently linked to the hydrogel by reverse transcription for subsequent whole-transcriptome amplification (WTA) using PCR (see Extended Data Fig. 1a–e). b,c, Hydrogel beads re-encapsulated on the second device are analysed using droplet PCR for HIV gag (b) and then sorted on the third device (c). d, The processing steps performed after droplet sorting include WTA, library preparation and sequencing, and bioinformatic comparison of HIV-DNA+ cells and HIV-DNA− cells. e, Droplet cytometry plots demonstrating the analysis of pure HIV-DNA+ J-Lat T cells (left), pure HIV-DNA− Jurkat T cells (right), and a mixture of 0.1% J-Lat and 99.9% Jurkat cells (middle). Cells were encapsulated at 1 cell per 10 droplets.” Reproduced under a Creative Commons Attribution 4.0 International License from Clark, I.C., Mudvari, P., Thaploo, S. et al. HIV silencing and cell survival signatures in infected T cell reservoirs. Nature (2023).
Figures and the abstract are reproduced from Clark, I.C., Mudvari, P., Thaploo, S. et al. HIV silencing and cell survival signatures in infected T cell reservoirs. Nature (2023). https://doi.org/10.1038/s41586-022-05556-6 under a Creative Commons Attribution 4.0 International License.
Read the original article: HIV silencing and cell survival signatures in infected T cell reservoirs


