21 Aug Microfluidic chemostat facilitates designing cell-free genetic networks
Synthetic biology and microfluidics have each brought enabling and revolutionary technological advances over the past few decades. However, they have advanced separately and their combined synergy has not yet been fully explored. For this week’s research highlight, we selected a recent Nature Communications paper in which a PDMS-based microfluidic chip was employed for designing cell-free genetic networks. The microfluidic chip served s a chemostat and provided a controlled environment for characterizing libraries of genetic building blocks followed and devising cell-free protein synthesis experiments.
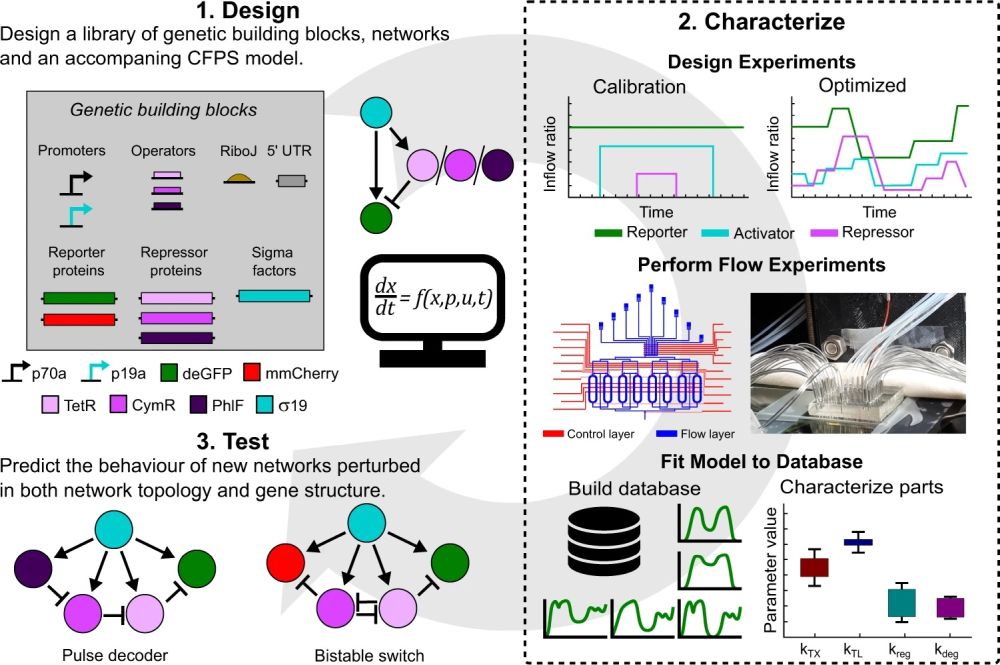
“Our methodology is based on a design–characterize–test cycle (Fig. 1), in which we combine microfluidics, optimal experimental design (OED) and optimize a kinetic model of the CFPS process with an agent-based non-linear least-squares optimization routine which utilizes all collected experimental data simultaneously “, the authors explained.

(1) IFFLs are assembled from a library of genetic building blocks. All IFFLs share the same activator but differ in the repressors. Besides, a CFPS ODE model is formulated to describe all reactions (Eqs. 3–9 in the Supplementary Information). (2) Calibration and optimized inflow patterns are created (top) and used to perform CFPS experiments in microfluidic chemostats (middle). Schematic representation of the microfluidic device (left) and a picture of a single device plugged in and ready for an experiment (right) are shown. All experiments are combined in a database and we estimate the parameter values and distributions using the associated kinetic models. (3) Using the same library of building blocks, two new networks are assembled and are used as test cases to assess the predictive power of the estimated parameters and model. General overview of the network topology is shown. ” Reproduced under Creative Commons Attribution 4.0 International License from van Sluijs, B., Maas, R.J.M., van der Linden, A.J. et al. A microfluidic optimal experimental design platform for forward design of cell-free genetic networks. Nat Commun 13, 3626 (2022).
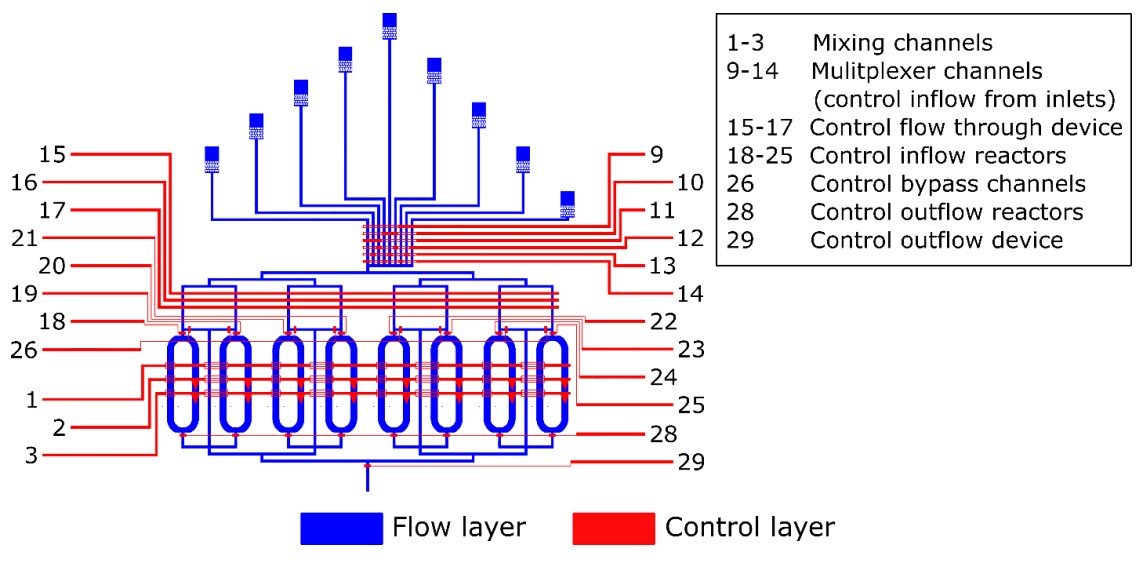
Layout of the microfluidic device based on the devices from Niederholtmeyer et al. and Van der Linden et al. The devices consist of two distinct layers, a flow layer (blue) and a control layer (red). At intersections of the two, pressurizing the control layer result in blocking the flow in the flow layer. The control layers are numbered, the purpose of each channel is provided in the legend. ” Reproduced under Creative Commons Attribution 4.0 International License from van Sluijs, B., Maas, R.J.M., van der Linden, A.J. et al. A microfluidic optimal experimental design platform for forward design of cell-free genetic networks. Nat Commun 13, 3626 (2022).
“In conclusion, we demonstrated a microfluidic platform coupled with a computational OED workflow capable of characterizing genetic building blocks for the modular construction of synthetic gene networks in CFPS systems. With this platform in place, future work will include an increase of the library of well-characterised modular building blocks and forward design of larger and more complex (cell-free) genetic networks.“, the authors explained.
Figures and the abstract are reproduced from van Sluijs, B., Maas, R.J.M., van der Linden, A.J. et al. A microfluidic optimal experimental design platform for forward design of cell-free genetic networks. Nat Commun 13, 3626 (2022). https://doi.org/10.1038/s41467-022-31306-3
Read the original article: A microfluidic optimal experimental design platform for forward design of cell-free genetic networks


