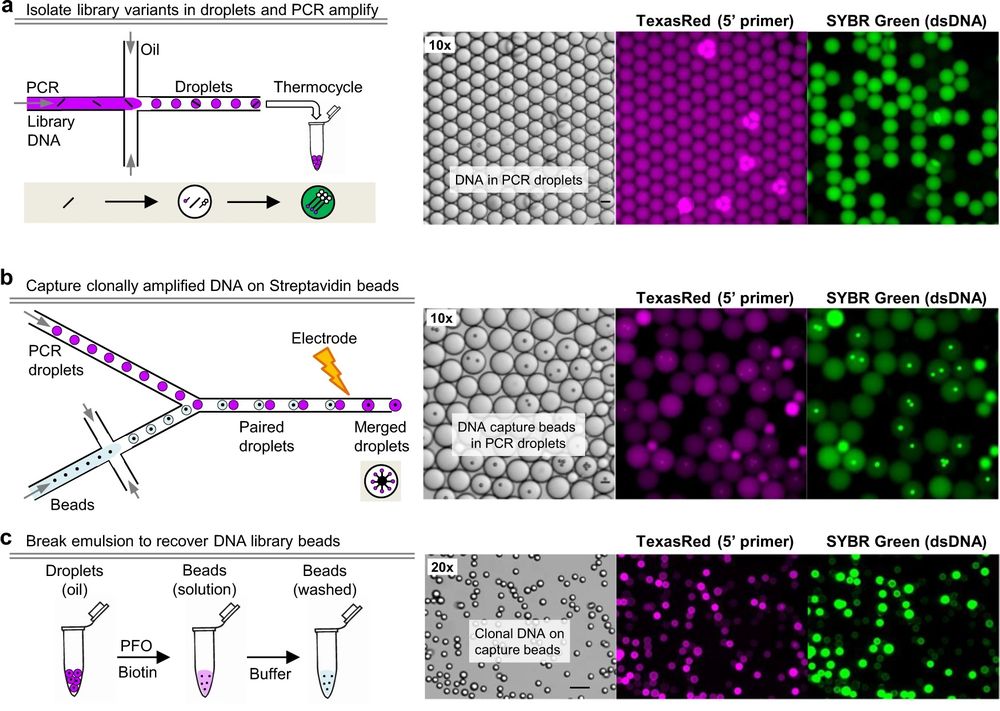
26 May A droplet microfluidic approach for generating genetically encoded fluorescent biosensors
Abstract
“Genetically encoded fluorescent biosensors are powerful tools used to track chemical processes in intact biological systems. However, the development and optimization of biosensors remains a challenging and labor-intensive process, primarily due to technical limitations of methods for screening candidate biosensors. Here we describe a screening modality that combines droplet microfluidics and automated fluorescence imaging to provide an order of magnitude increase in screening throughput. Moreover, unlike current techniques that are limited to screening for a single biosensor feature at a time (e.g. brightness), our method enables evaluation of multiple features (e.g. contrast, affinity, specificity) in parallel. Because biosensor features can covary, this capability is essential for rapid optimization. We use this system to generate a high-performance biosensor for lactate that can be used to quantify intracellular lactate concentrations. This biosensor, named LiLac, constitutes a significant advance in metabolite sensing and demonstrates the power of our screening approach.”
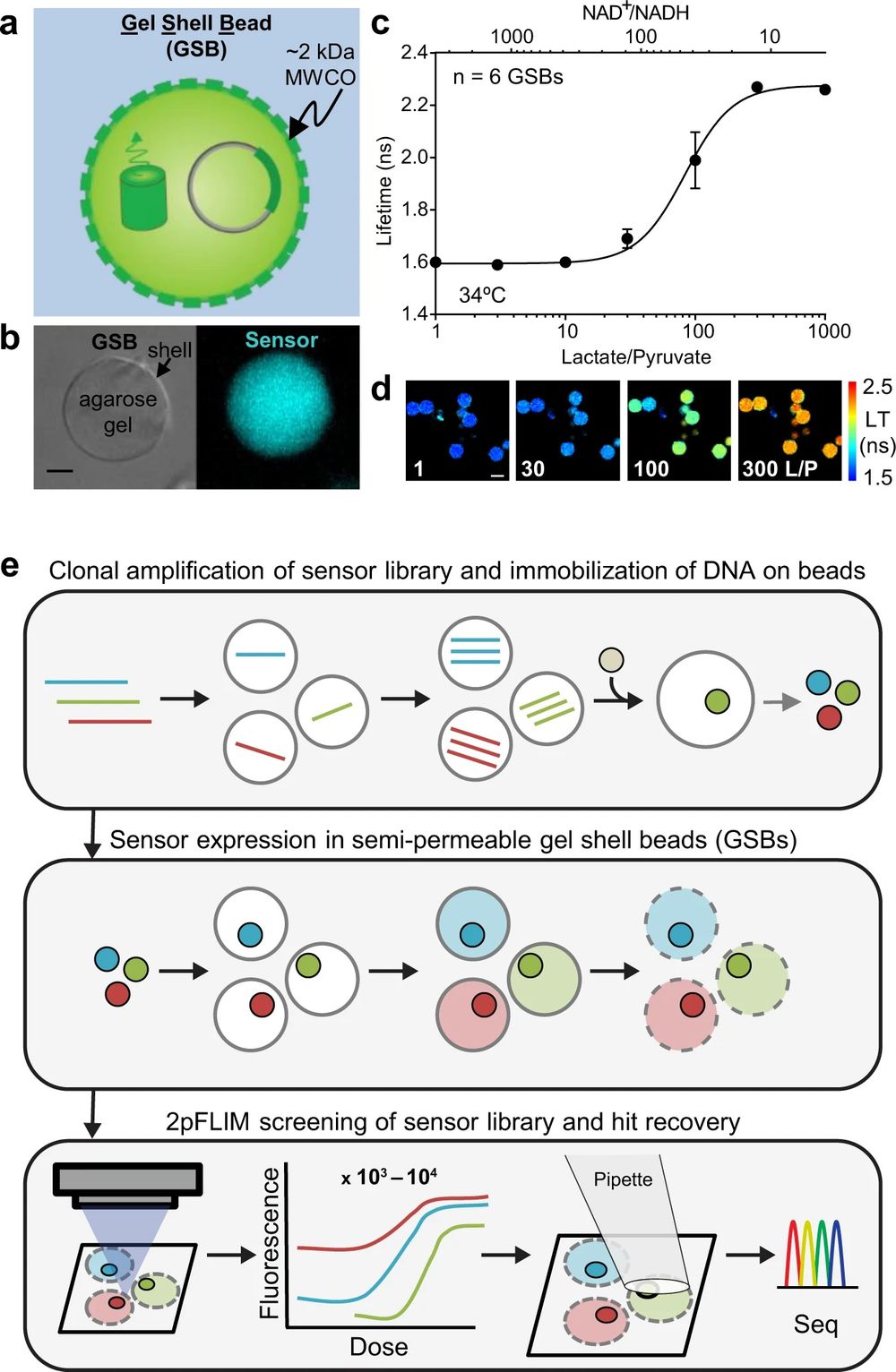
“a Cartoon depiction of a GSB containing plasmid DNA and fluorescent biosensor protein. An agarose gel core is wrapped in a polyelectrolyte shell with a reported molecular weight cutoff of ~2 kDa. b A single 20 μm GSB doped with a green fluorescent biosensor, brightfield (left) and epifluorescence (right), 5 μm scale bar. c Dose response curve of Peredox, a fluorescence lifetime sensor that responds to NAD+/NADH, acquired at pH 7.3 at 34 °C on purified biosensor protein encapsulated in GSBs (mean ± SD). Purified lactate dehydrogenase (LDH) plus lactate and pyruvate was used to set the NAD+/NADH ratio outside the GSBs, following methods previously established for Peredox. LDH does not enter the GSBs. d Filmstrip of the fluorescence lifetime images used to generate the data in (c), pseudocolored according to the lifetime heatmap at the right, and arranged in order of Lactate/Pyruvate (L/P). 30 μm scale bar. e BeadScan overview schematic. Single plasmids from a biosensor library are isolated in microfluidic droplets that serve as picoliter-sized reaction chambers for PCR-based amplification. Clonal amplicons are immobilized on affinity beads introduced into each droplet, ultimately yielding a library of clonal DNA beads. Single beads are then re-encapsulated in new droplets containing in vitro transcription/translation (IVTT) reagents for biosensor expression. IVTT droplets containing mature biosensor are then converted into semipermeable gel-shell beads (GSBs). Up to 10,000 GSBs are arrayed on a glass coverslip for automated fluorescence imaging and analysis. Single GSBs displaying favorable biosensor properties are collected via micropipette, and the genotypes recovered from the DNA bead by PCR and Sanger sequencing.” Reproduced under Creative Commons Attribution 4.0 International License from Koveal, D., Rosen, P.C., Meyer, D.J. et al. A high-throughput multiparameter screen for accelerated development and optimization of soluble genetically encoded fluorescent biosensors. Nat Commun 13, 2919 (2022).
Figures and the abstract are reproduced from Koveal, D., Rosen, P.C., Meyer, D.J. et al. A high-throughput multiparameter screen for accelerated development and optimization of soluble genetically encoded fluorescent biosensors. Nat Commun 13, 2919 (2022). https://doi.org/10.1038/s41467-022-30685-x. under Creative Commons Attribution 4.0 International License.
Read the original article: A high-throughput multiparameter screen for accelerated development and optimization of soluble genetically encoded fluorescent biosensors


