21 Apr Mechanotyping of individual cells using deep-learning assisted microfluidics
Abstract
“Mechanical properties of cells are important features that are tightly regulated and are dictated by various pathologies. Deformability cytometry allows for the characterization of the mechanical properties at a rate of hundreds of cells per second, opening the way to differentiating cells via mechanotyping. A remaining challenge for detecting and classifying rare sub-populations is the creation of a combined experimental and analysis protocol that approaches the maximum potential classification accuracy for single cells. In order to find this maximum accuracy, we designed a microfluidic channel that subjects each cell to repeated deformations and relaxations and provides a comprehensive set of mechanotyping parameters. We track the shape dynamics of individual cells with high time resolution and apply sequence-based deep learning models for feature extraction. In order to create a dataset based solely on differing mechanical properties, a model system was created with treated and untreated HL60 cells. Treated cells were exposed to chemical agents that perturb either the actin or microtubule networks. Multiple recurrent and convolutional neural network architectures were trained using time sequences of cell shapes and were found to achieve high classification accuracy based on cytoskeletal properties alone. The best model classified two of the sub-populations of HL60 cells with an accuracy over 90%, significantly higher than the 75% we achieved with traditional methods. This increase in accuracy corresponds to a fivefold increase in potential enrichment of a sample for a target population. This work establishes the application of sequence-based deep learning models to dynamic deformability cytometry.”

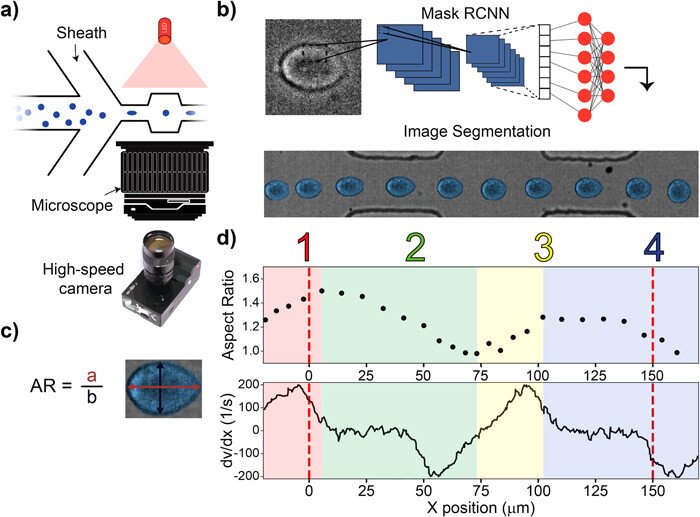
Principles of repeated mechanotyping. (a) Channel design utilizing sheath flow, a high-powered LED, and a microscope. The microfluidic channel that enables characterization and classification contains a cavity placed between two narrow zones. (b) Data are captured by a high-speed camera, creating videos at 11 k fps. Cell borders are detected and fit using Mask-RCNN. (c) The cell deformation, AR, was quantified as the ratio of two axes of an ellipse that approximates the cell’s shape. (d) (Top) The aspect ratio vs position relative to channel entrance of a single cell as it passes through the channel. (Bottom) COMSOL simulation showing the derivative of velocity vs channel position, which is proportional to the shear stress. Region 1 (R1) and Region 3 (R3), denoted by red and yellow regions, are where the cells undergo deformation. Region 2 (R2) and Region 4 (R4), denoted by green and blue regions, are where the cells undergo relaxation. Reproduced under Creative Commons Attribution 4.0 International License from Cody Combs, Daniel D. Seith, Matthew J. Bovyn, Steven P. Gross, Xiaohui Xie, and Zuzanna S. Siwy, “Deep learning assisted mechanotyping of individual cells through repeated deformations and relaxations in undulating channels”, Biomicrofluidics 16, 014104 (2022).
Figures and the abstract are reproduced from Cody Combs, Daniel D. Seith, Matthew J. Bovyn, Steven P. Gross, Xiaohui Xie, and Zuzanna S. Siwy , “Deep learning assisted mechanotyping of individual cells through repeated deformations and relaxations in undulating channels”, Biomicrofluidics 16, 014104 (2022) https://doi.org/10.1063/5.0077432 under Creative Commons Attribution 4.0 International License.
Read the original article: Deep learning assisted mechanotyping of individual cells through repeated deformations and relaxations in undulating channels featured


