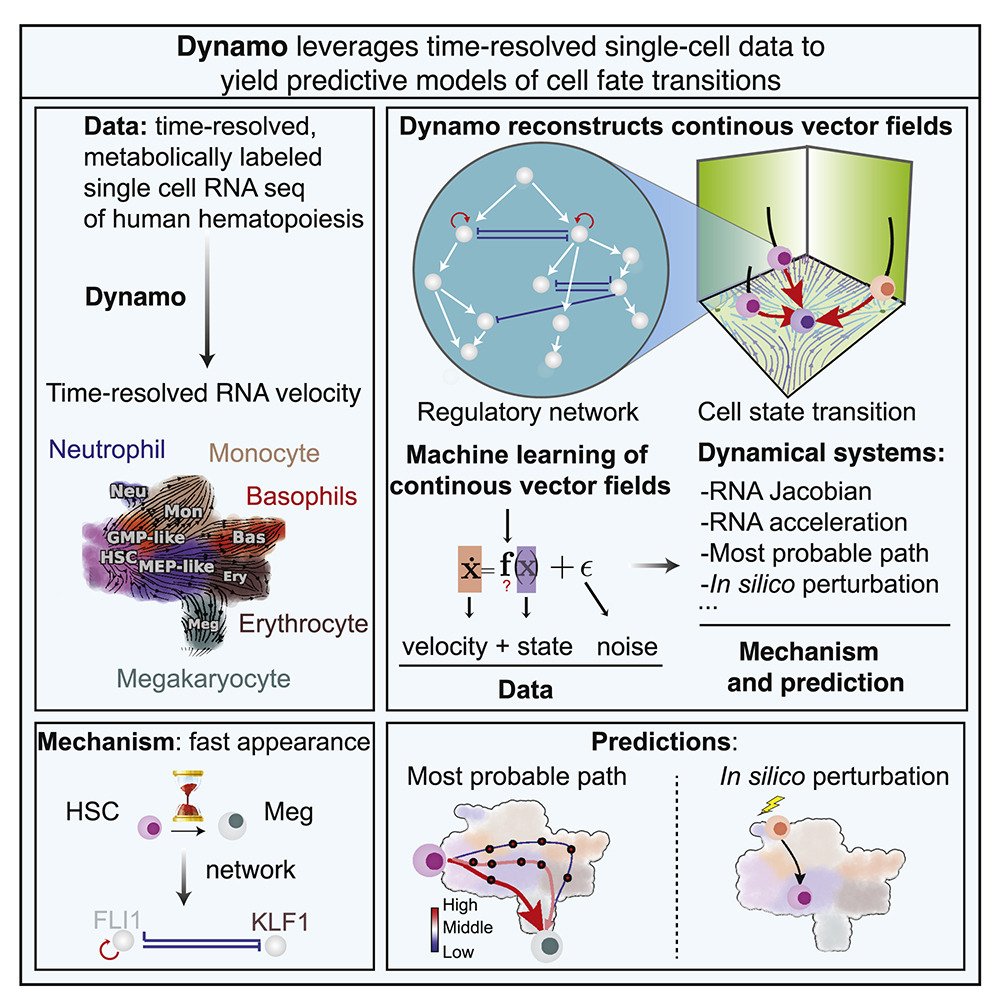
19 Mar Cellular transcriptomic vector fields revealed using dropseq microfluidic chips
Understanding the dynamic transitions of cell states has been a formidable challenge in cellular biology. Traditional methods have struggled to capture these intricate processes at a single-cell level. A recent study published in Cell introduces a new approach that leverages microfluidic technology to map transcriptomic vector fields of single cells, providing a groundbreaking solution to this complex challenge. The study employs advanced dropseq microfluidic chips microfabricated at uFluidix to facilitate the detailed analysis of single-cell RNA sequencing (scRNA-seq) data. The integration of our advanced microfluidic microfabrication techniques in microfluidic devices enables precise manipulation and analysis of individual cells, enhancing the accuracy and depth of biological insights. Dynamo, the computational framework developed in this research, reconstructs continuous transcriptomic vector fields from discrete RNA velocity vectors. It offers a comprehensive view of cell fate, regulatory mechanisms, and cell-state transitions, shedding light on the dynamics of gene regulatory networks.

Reproduced under Creative Commons Attribution 4.0 International License from Qiu, X., Zhang. Y., et al. Mapping transcriptomic vector fields of single cells. Cell 185, 690-711 (2022).
Dynamo, the newly developed analytical tool, reconstructs continuous transcriptomic vector fields from discrete RNA velocity vectors. It provides a comprehensive analysis of cell fate, regulatory mechanisms, and cell-state transitions based on scRNA-seq data. Dynamo integrates RNA metabolic labeling with expression kinetics to estimate RNA kinetic parameters across genome-wide data. The framework overcomes limitations of conventional splicing-based RNA velocity analyses, enabling accurate estimation of RNA turnover rates and cellular dynamics.
Dynamo’s differential geometry analyses revealed the mechanisms driving early megakaryocyte appearance and asymmetrical regulation within the PU.1-GATA1 circuit. Using the least action path method, Dynamo accurately predicts drivers of numerous hematopoietic transitions. In silico perturbations can predict cell fate diversions induced by gene perturbations, offering new insights into cellular behavior.
The study’s findings underscore the potential of transcriptomic vector fields in elucidating the dynamics of gene regulatory networks. The ability to predict cell fate and the outcomes of genetic perturbations opens up new possibilities for biomedical research and therapeutic strategies. This innovative use of microfluidics in transcriptomic analysis opens up new avenues for understanding cellular behavior and gene regulation. The predictive capabilities of Dynamo in determining cell fate have far-reaching implications in fields ranging from developmental biology to medical therapeutics. The study marks a significant advancement in single-cell genomics, blending microfluidics with computational analysis to bring a quantitative and predictive understanding of cell-state transitions. This approach paves the way for a deeper exploration of the complexities of life at the cellular level.
Figures and the abstract are reproduced from Qiu, X., Zhang. Y., et al. Mapping transcriptomic vector fields of single cells. Cell 185, 690-711 (2022). https://doi.org/10.1016/j.cell.2021.12.045 under Creative Commons Attribution 4.0 International License
Read the original article: Mapping transcriptomic vector fields of single cells


