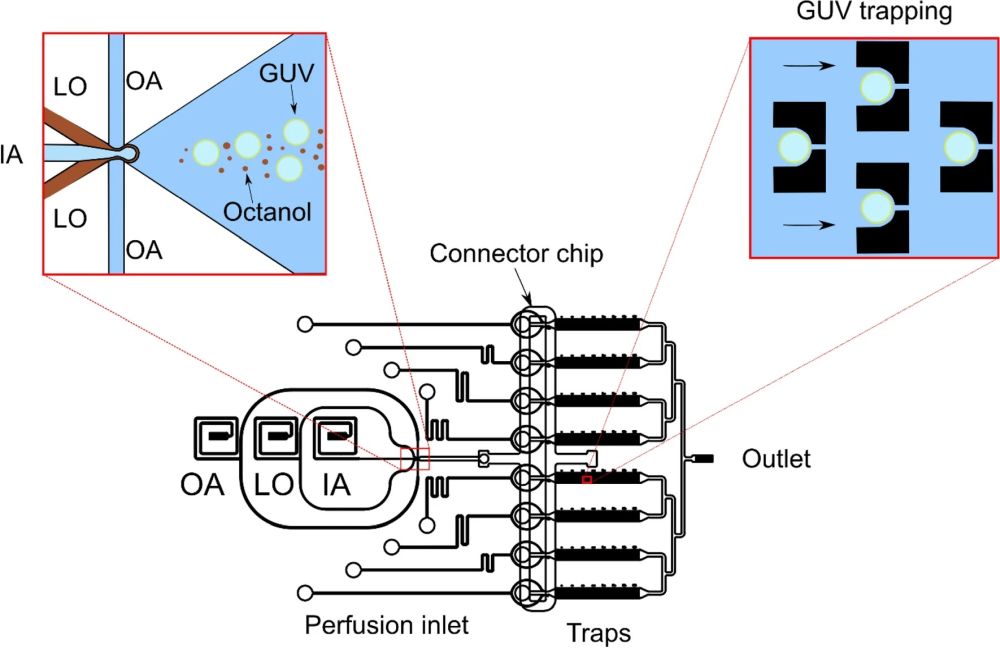
07 Mar Microfluidic analysis of peptides reveals a correlation between physio-chemical properties and biological activity
“Antimicrobial resistance challenges the ability of modern medicine to contain infections. Given the dire need for new antimicrobials, polypeptide antibiotics hold particular promise. These agents hit multiple targets in bacteria starting with their most exposed regions—their membranes. However, suitable approaches to quantify the efficacy of polypeptide antibiotics at the membrane and cellular level have been lacking. Here, we employ two complementary microfluidic platforms to probe the structure–activity relationships of two experimental series of polypeptide antibiotics. We reveal strong correlations between each peptide’s physicochemical activity at the membrane level and biological activity at the cellular level. We achieve this knowledge by assaying the membranolytic activities of the compounds on hundreds of individual giant lipid vesicles, and by quantifying phenotypic responses within clonal bacterial populations with single-cell resolution. Our strategy proved capable of detecting differential responses for peptides with single amino acid substitutions between them, and can accelerate the rational design and development of peptide antimicrobials.”
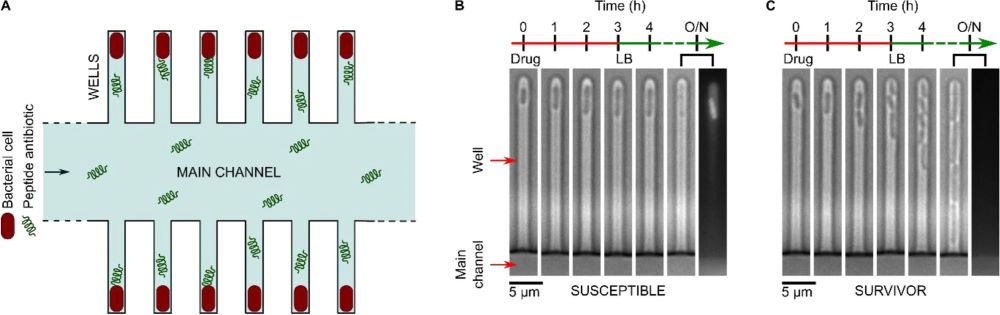
“Probing the biological activity of experimental polypeptide antibiotics with single-cell resolution. The mother-machine microfluidic device, whose schematic is shown in (A), consists of a “Main channel” for seeding the connected side channels or “wells” of the device with cells. Drugs, nutrients, and viability stains are then dosed to the trapped cells via the main channel. In the microscopy images tracking individual wells in (B) and (C) above, we observe examples of individual susceptible (B) and survivor (C) cells in response to peptide treatment in the mother-machine microfluidic device. The two examples above are taken from the same experiment and show the contrasting responses of two clonal E. coli cells to the peptide bienK11 (10 µM). The cell shown in (B) was susceptible to the treatment, halting growth during the 3 h of drug dosage and disintegrating thereafter when the drug was replaced with fresh Lysogeny Broth (LB) and incubated overnight (O/N). The disintegration is clearly visible when comparing the 4 h and O/N time-points—in the final fluorescence image panel, taken after O/N LB treatment, we see the debris of the cell stained with the dead stain propidium iodide (PI). In contrast, the cell shown in (C) resisted the treatment, growing and dividing even during peptide delivery, with no PI staining in the daughter cells at the end of the experiment.”. Reproduced under Creative Commons Attribution 4.0 International License Cama, J., Al Nahas, K., Fletcher, M. et al. An ultrasensitive microfluidic approach reveals correlations between the physico-chemical and biological activity of experimental peptide antibiotics. Sci Rep 12, 4005 (2022).
Figures and the abstract are reproduced from Cama, J., Al Nahas, K., Fletcher, M. et al. An ultrasensitive microfluidic approach reveals correlations between the physico-chemical and biological activity of experimental peptide antibiotics. Sci Rep 12, 4005 (2022). https://doi.org/10.1038/s41598-022-07973-z under Creative Commons Attribution 4.0 International License
Read the original article: An ultrasensitive microfluidic approach reveals correlations between the physico-chemical and biological activity of experimental peptide antibiotics


