05 Jan Double emulsions for analyzing gene expression dynamics in cell-free systems
Welcome back, science enthusiasts! Today, we’re delving into the fascinating world of synthetic cell populations and exploring a groundbreaking study that unraveled the intricate dynamics of gene expression within these miniature artificial cells. This remarkable research sheds light on the behavior of cell-free expression systems (CFES) and opens up exciting possibilities for synthetic biology and biotechnology.
In the realm of synthetic biology, researchers are harnessing the power of microfluidics to unravel the mysteries of gene expression dynamics within synthetic cell populations. Leveraging microfluidic chips and ingenious microfabrication techniques, scientists are paving the way for groundbreaking advancements in biotechnology and medical research. In this blog post, we’ll delve into the captivating study titled “Cell-Free Gene Expression Dynamics in Synthetic Cell Populations” to explore the role of microfluidics in understanding gene expression processes at the microscale.
Microfluidics is a transformative technology that allows precise control and manipulation of tiny amounts of fluids on a microscale. This technology has revolutionized various areas of biological research by enabling high-throughput experiments and providing a deeper understanding of cellular processes. In this study, microfluidics played a pivotal role in creating synthetic cell populations within tiny lipid-based compartments, resembling artificial cells, to investigate gene expression dynamics.
Synthetic cell populations offer a remarkable opportunity to mimic living organisms in a controlled environment. Crafted with precision, these artificial cells create microscale ecosystems that faithfully replicate cellular processes. The study in question designed a plasmid housing red fluorescent mCherry protein and Broccoli RNA aptamers, enabling simultaneous monitoring of mRNA transcription and protein translation.
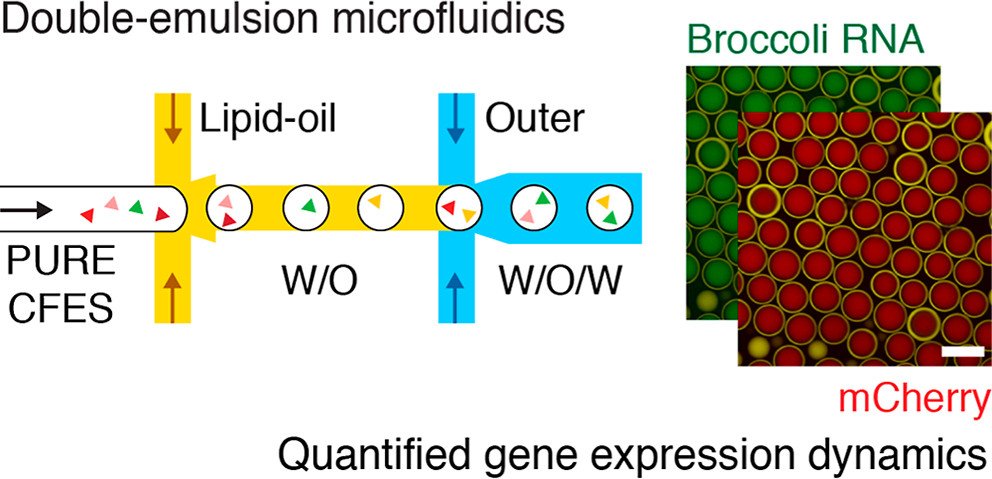
To investigate gene expression dynamics within the synthetic cell populations, the researchers utilized a unique microfluidic device that allows real-time monitoring of RNA and protein levels. This innovative approach enabled them to study both transcription and translation processes simultaneously, providing valuable insights into the intricate interplay between these fundamental molecular processes. Harnessing the power of microfluidics, the researchers encapsulated cell-free expression systems (CFES) in synthetic cells using a double-emulsion microfluidic device. This ingenious technique produced highly uniform synthetic cell populations, allowing the investigation of gene expression dynamics within a precisely controlled environment. The microfluidic chip played a pivotal role in this breakthrough, enabling precise control over the encapsulation process and promoting efficient mixing of reactants within the synthetic cells..
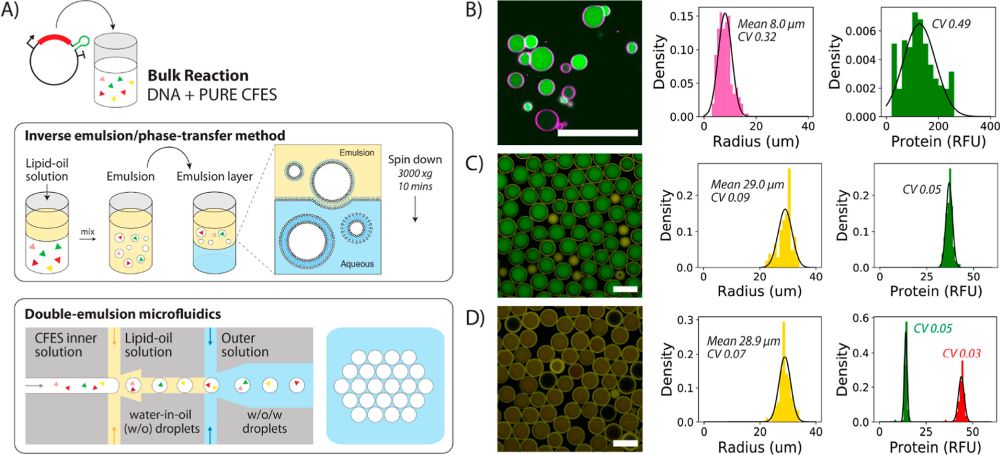
“Variability in synthetic cell populations. (A) Schematic of the bulk inverse emulsion phase transfer method and double-emulsion microfluidics to generate liposomes or synthetic cells. (B) Synthetic cell population expressing eGFP protein from 1.17 nM pEXP5-NT/6xHis eGFP plasmid DNA generated using the bulk inverse emulsion phase transfer method. (C) Microfluidic-generated synthetic cells expressing eGFP protein from 4.5 nM pEXP5-NT/6xHis eGFP F30-2xdBroccoli plasmid DNA. (D) Merged image of the synthetic cell population expressing both eGFP and mCherry protein from two plasmids (4.5 nM pEXP5-NT/6xHis eGFP and 4.5 nM pEXP5-NT/6xHis mCherry plasmid DNA). Endpoint histograms of radius and protein RFU are plotted alongside each of the synthetic cell populations (B–D). The number of cells analyzed is 206, 106, and 85 for (B–D), respectively. Black lines are Gaussian distributions obtained by fitting mean and variance of the data. These experiments show the relative levels of expressed protein and do not refer to absolute concentrations. RFU values between the microfluidic-generated synthetic cells in (C,D), but not the inverse emulsion-made synthetic cells, are comparable, as these images were acquired using the same microscopy settings. However, CV values can be compared across all populations. All images are taken at the endpoint after 12 h of incubation at 30 °C using confocal microscopy with a 40× objective for (B) and 10× objective for (C,D). Scale bars are all 100 μm.” Reproduced under Creative Commons Attribution 4.0 International License from Gonzales, T, D., Yandarpalli, N., Robinson, T., Zechner, C., Tang, D, T-Y., Cell-free Gene Expression Dynamics in Synthetic Cell Populations. Acs. Syn. Bio. 2022: https://doi.org/10.1021/acssynbio.1c00376
The researchers first constructed a plasmid, pEXP5-NT/6xHis mCherry F30-2xdBroccoli, consisting of a constitutive T7 RNA polymerase-mediated promoter to express a red fluorescent mCherry protein and two copies of a dimeric Broccoli RNA aptamer. The Broccoli RNA aptamer, upon binding to a small-molecule dye, emits a green fluorescence signal. This setup allowed simultaneous fluorescence monitoring of transcribed mRNA and reporter protein levels.
Bulk CFES experiments were conducted using a standard half-volume reaction mix of the PURExpress in vitro protein synthesis kit, supplemented with sucrose for osmolarity balance. The reaction mixtures were titrated with the pEXP5-NT/6xHis mCherry F30-2xdBroccoli plasmid DNA or its purified mRNA transcripts. Fluorescence measurements were taken at 10-minute intervals for 8 hours, and relative fluorescence units were converted into concentration units using calibration curves.
For the microfluidic experiments, synthetic cell populations were generated using a double-emulsion microfluidic device. The inner CFES solutions were prepared with varying plasmid DNA concentrations, and the lipid oil phase contained fluorescent dye DiD for imaging. The outer aqueous solution contained all the necessary components for gene expression. The synthetic cell populations were imaged by confocal microscopy at 30 °C for 12 hours.
Accompanying the microfluidic experiments was the development of a novel coarse-grained mathematical model based on resource-limited gene expression dynamics. This model allowed researchers to quantitatively compare and estimate gene expression parameters without requiring complete knowledge of the CFES composition. It provided valuable insights into the kinetics of transcription and translation, unraveling the temporal dynamics of gene expression within the synthetic cell populations.
The seamless integration of microfluidics and mathematical modeling has propelled the field of synthetic biology to new heights. This research not only advances our fundamental understanding of gene expression but also holds great potential for applications in biotechnology, medicine, and beyond.
Figures and the abstract are reproduced from N., Robinson, T., Zechner, C., Tang, D, T-Y., Cell-free Gene Expression Dynamics in Synthetic Cell Populations. Acs. Syn. Bio. 2022: https://doi.org/10.1021/acssynbio.1c00376 under Creative Commons Attribution 4.0 International License
Read the original article: Cell-Free Gene Expression Dynamics in Synthetic Cell Populations


