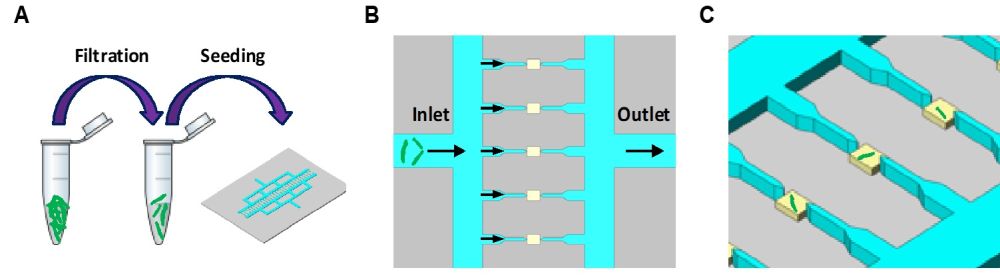
07 Jun Microfluidic chip for growth visualization of Mycobacterium smegmatis at single-cell resolution
“Here, we combine an acoustically driven manipulation device with a micro-force sensor to freely rotate biological samples and quantify mechanical properties at multiple regions of interest within a specimen. The versatility of this tool is demonstrated through the analysis of single Lilium longiflorum pollen grains, in combination with numerical simulations, and individual Caenorhabditis elegans nematodes.”, the authors explained.
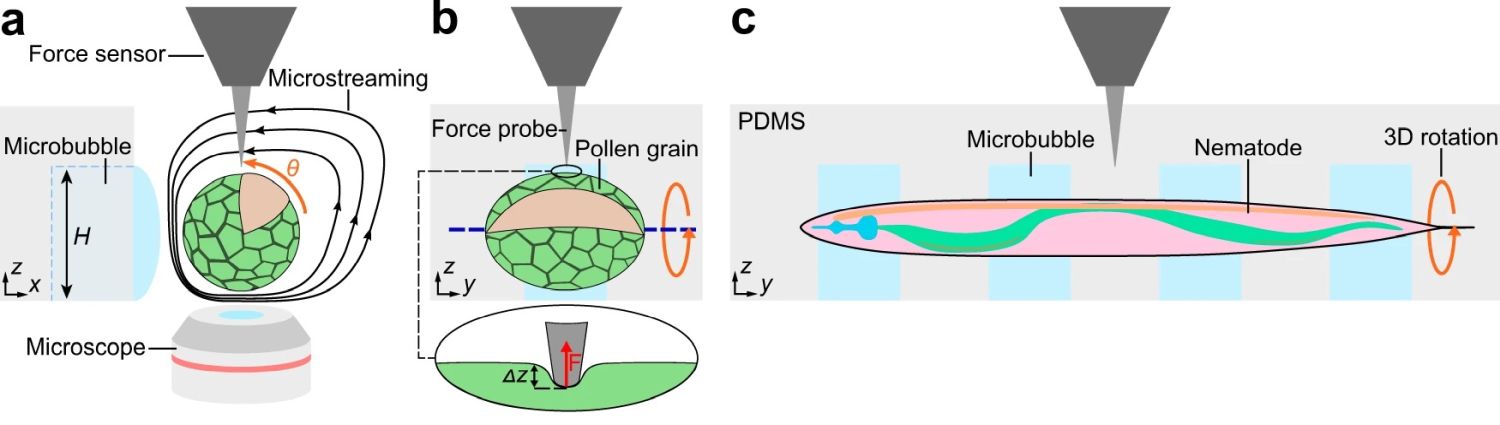
Reproduced under Creative Commons Attribution 4.0 International License. Läubli et al., Nat. Commun., 2021.
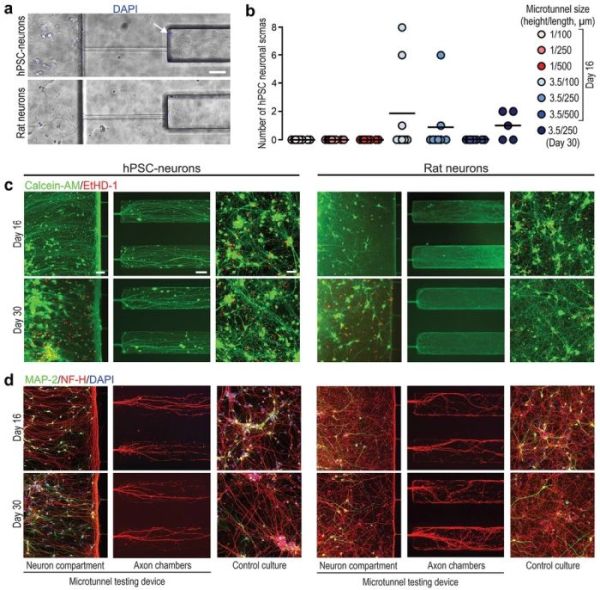
Reproduced under Creative Commons Attribution 4.0 International License. Ristola et al., Adv. Mater. Interfaces., 2021.
“In this study, we showed that successful isolation of axons from somas and dendrites combined with the robust axonal outgrowth of hPSC-derived and rat primary cortical neurons in a compartmentalized microfluidic PDMS device is greatly influenced by the cross-sectional area and the length of the separating microtunnels. Furthermore, we created an hPSC-derived neuron-based axonal model consisting of a compartmentalized PDMS microtunnel chip integrated with photoinscribed nanotopography on light-responsive, azobenzene-based molecular glass.”, the authors concluded.
Read the original article: Directional Growth of Human Neuronal Axons in a Microfluidic Device with Nanotopography on Azobenzene‐Based Material
Pouriya Bayat
Pouriya is a microfluidic production engineer at uFluidix. He received his B.Sc. and M.A.Sc. both in Mechanical Engineering from Isfahan University of Technology and York University, respectively. During his master’s studies, he had the chance to learn the foundations of microfluidic technology at ACUTE Lab where he focused on designing microfluidic platforms for cell washing and isolation. Upon graduation, he joined uFluidix to even further enjoy designing, manufacturing, and experimenting with microfluidic chips. In his free time, you might find him reading a psychology/philosophy/fantasy book while refilling his coffee every half an hour. Is there a must-read book in your mind, do not hesitate to hit him up with your to-read list.


