Microfluidic approaches for performing microarrays assays and microarray patterning
Microarrays are powerful tools in molecular biology. They allow multiplex and high-throughput analysis of biological analytes. They consist of an array of biological substances allowing multiple tests to be done simultaneously. Biomolecules (such as DNA and protein) and cellular microarrays are common among researchers.
DNA microarray is a standard technique for analyzing gene expression levels. They include an array of microscopic spots that react with the genetic material in a sample to form a double DNA strand. Conventionally, the control and experimental samples should sit on the microarray for incubation. In this manner, the main means of mass transport is transportation by diffusion.
Protein microarrays, on the other hand, are powerful tools for diagnostics and proteomics. They include an array of capture proteins that interact with the proteins in the sample and normally emit fluorescence signal that can be read by a scanner.
Cell microarrays are systems of fixed or living cells that are immobilized on a solid substrate and are strong tools for screening multiple cells at the same time.
Microfluidics, however, is associated with controlling very small volumes of liquid within microchannels. Microfluidic technology can enhance exposure by running the samples over the microarrays. Additionally, microfluidic chips are excellent choices for printing novel low-density and high-density arrays.
How does microfluidic technology improve microarray assays?
Microfluidic chips’ dimensions normally range from several microns to a couple of hundred microns. These micron-sized microfluidic channels can be filled with very small amounts of the samples. Therefore, in microfluidic experiments, the sample volume is highly reduced compared to conventional and bulk methods. This sample reduction reduces the cost of the reagents and the experiment.
In a typical microarray test, the sample is placed onto the microarray slide where it rests for hours for incubation. The hybridization is dominated by diffusion which is not efficient and is very time-consuming. However, microfluidic resolves this issue by flowing the reagents over the hybridization spots using various methods such as capillary flow, electrokinetic techniques, or syringe/pressure pumps. Direct and rapid exposure of the spots to the reagents dramatically reduces the hybridization time and increases the efficiency.
Moreover, in many cases, multiple tests need to be done in parallel. Microfluidic devices can be easily paralleled to allow multiple samples to be tested at once or to allow each sample to be tested multiple times to ensure the validity of the results.
DNA microarrays
DNA microarrays consist of an array of DNA probes attached to a solid substrate. Each of the spots hosts a target gene that can bind to its counterpart in the sample. The sample is usually equipped with fluorescent probes. Thus, if a gene in the sample pairs with a particular gene the associated spot will fluoresce. The researcher can then analyze the microarray using a computer to measure the expression levels. Microfluidic chips have been used both for DNA microarray assays and patterning novel DNA probes.

Microfluidic microarray assays
Commercial or home-made microarrays are often printed on glass and microfluidic chips easily bond to the glass. A common approach for performing microfluidic microarray tests is by bonding the glass microarray to the microfluidic chip. The microarray can be aligned with microchannels in a microfluidic device such that after the bonding running the sample through the microchannels would expose the genes to the DNA spots. Large surface to volume ratio of microchannels increases the chance of pairing genes with the DNA probes. In microfluidic microarray assays, the reagents are not stationary and flow over the microarray spots. It is this motion, unlike conventional techniques, that increase efficiency and reduces hybridization time dramatically. For increasing efficiency, the sample can recirculate or oscillate back and forth over the region of interest. Paralleling the chips increases the throughput and multiplexing. Several microarrays with the same structure can sit in parallel to test the repeatability of the assay. Also, different samples such as control and experimental sample can run in parallel over the same microarray structure.
Various types of microfluidic devices can be fabricated for this purpose. Depending on the experiment and the microarray type, both straight and spiral microchannels can be used in microarray tests. Using these microchannels removes the need for large hybridization chambers that reduces the sample volume and cost and increases efficiency.
Microfluidics fluid delivery to the microarray chips
Depending on the type of the experiment, the reagents, and the microarray density, the fluid delivery mechanism can differ. Electroosmotic flow requires a particular type of sample that makes its application limited. Also, it can change the properties of the fluids that in turn can affect the results. Syringe pumps and pressure pumps are very common and precise means of handling the fluids. However, in microarray applications, the number of ports might exceed the number of available connections in a syringe pump or a pressure pump rendering them inapplicable. Centrifugal microfluidics can overcome this issue. In centrifugal microfluidics, the sample is loaded in microfluidic chips placed on a rotor. The fluid will be delivered to the desired points due to the rotation of the microfluidics chip and the centrifugal force imposed on the reagents.
Microarray patterning using microfluidic chips
The quality of a microarray assay is highly dependent on the quality of the DNA spots. Regular low-density probe printing is based on pin-spotting machines. However, since the spotting reagent and the slide are both exposed to air, the technique suffers from splashing and evaporation. Moreover, spotting with this method can be heterogeneous whereas a homogenous probe results in more accurate image analysis. Microfluidics is specifically beneficial for patterning low-density microarrays. A simple microfluidic technique can be used to print line arrays and overcome the abovementioned issues. For this, a network of imprinted microchannels in a polymer (normally PDMS) reversibly bonds to a solid substrate. This bonding does not need to be as strong as irreversible bonding such as plasma bonding. Next, the solutions run through these channels and thus pattern the surface of the substrate. The microchannels can then be washed to clean the unreacted reagents. Since the microchannels are separate the risk of cross-contamination is reduced. Finally, the microchannel detaches and leaves the patterned substrate. In this approach, the reagents flow in a confined space and are not exposed to the air. Therefore, there is no risk of splashing or evaporation.

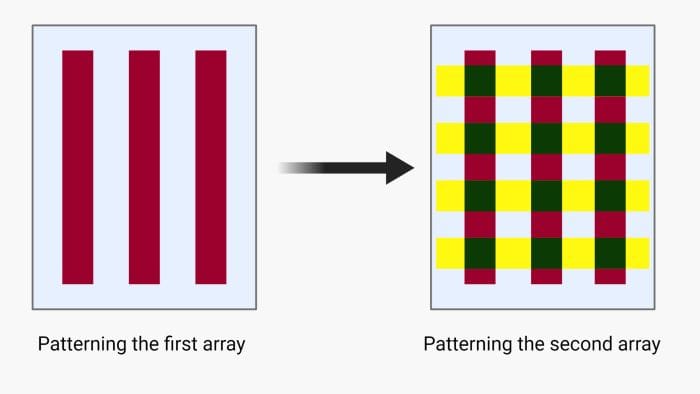
Another microfluidic technique for low-density microarray printing is called the intersection method or micromosaic method. Here, the first array is patterned similarly to the linear arrays explained above. Next, the microchannels will be unsealed and another chip will be place orthogonal to the first array and the same process for patterning will be followed. This method was first used for immunoassays and then was extended to DNA microarrays and can form rectangular patches on the solid substrate making it suitable for parallel hybridization. The intersection method can be done using two sets of orthogonal straight microchannels or spiral and radial microfluidics channels. In the latter, the first array forms circles (or a spiral) and is cross-sectioned by the second array of radial lines. This method is suitable for cases where centrifugal microfluidics is the method of choice for delivering the samples.
 More sophisticated methods pattern the probes in 3D matrices or employing beads rather than planar patterning. Microbeads have larger surface areas and are more effective in capturing the target DNAs. These beads normally are modified with a specific probe and trapped in microwells where the stream of the experimental sample flows over. Since beads are attached to different probes, they can be identified based on their location to prepare the genetic library.
More sophisticated methods pattern the probes in 3D matrices or employing beads rather than planar patterning. Microbeads have larger surface areas and are more effective in capturing the target DNAs. These beads normally are modified with a specific probe and trapped in microwells where the stream of the experimental sample flows over. Since beads are attached to different probes, they can be identified based on their location to prepare the genetic library.


